DNA Repair and Genome Integrity 1/2
1/26
Earn XP
Description and Tags
Lecture 8
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
27 Terms
Why is DNA subject to constant change?
it is getting damaged constantly because of all the reactions in the cell
exampled of the constant change to which DNA is subjected
hydrytic depurinations (water interactions in the genome)
2000-100000 /day/cell
cytosune deaminations
1/5days/cell
guadinine oxidaion
1/5days/cell
methylation of adenine
600Lday/cell
=> these account for a lot of change and DNA repari potential because there ares estimated 3.72×10^13 cells in our body
what are three processes that produce modified bases? and what can the modification of bases do ?
deamination
when amine group is removed => chemistry of base is changed
oxidation
alkylation
small molecules containing carbon are attacehd to these bases (i.e. metjhylation)
this can alter the original base pairring
may block the activity of the DNA polylerases during DNA replication
so have to be reparied for DNA replication to occur
mutations
permanent, transmissible, changed to the genetic material of a cell or organism
can occur spontaneously
can occur by transposable elements (jumoing genes)
can occur because of errors during replication
mutagens
chemical compounds like UV radiation or ionizing radiation that increase the frequency of mutations
carcinogen
an agent that causes cancer
many carcinogens are mutagens
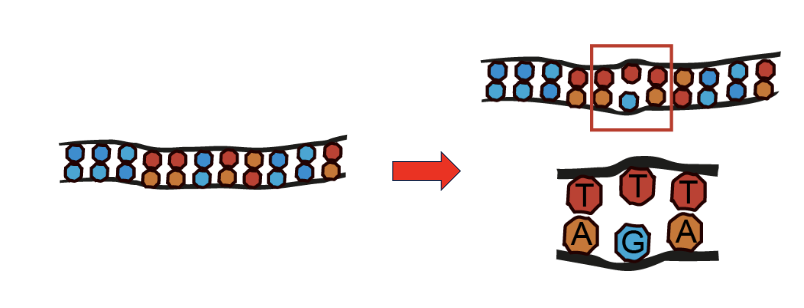
Where do Mismatches come from?
DNA polymerase can introduce errors
alone introduces 1 error every 10000 nucleotides
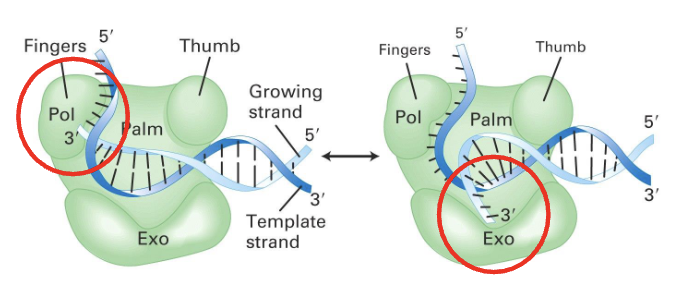
what are the repair mechanisms for mismatch errors?
proofreading exonuclease
an activity of DNA polymerases to correct the mistakes they made
activity increases fidelity by 100 fold
MMR: Mismatch repair
further increases fidelity by 1000 fold
adding this up: probability of incorporating a single mistake of 10^9 or 10^10 per nucleotide in the human cell
why does the n° of mutations fathers ingerit to their offspring increase with age?
because of continuous production of germ cells
what can we say about the rate of mutations in viruses vs cells
It is much higher in viruses
for higher probability of selecting more successful variants
what DNA polymerases have a 3’ to 5’ exonuclease activity?
DNA polymerase epsilon
DNA polymerase delta
(not DNA pol alpha!!)
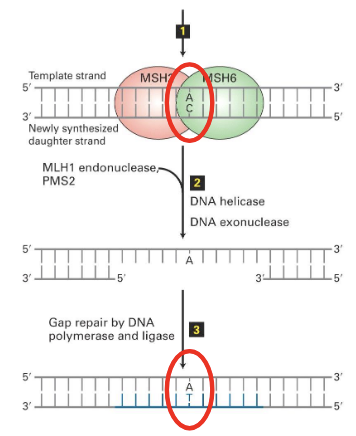
where does MMR often act in proximity to?
the replisome
with what protein from the replisome do some of the proteins from the MMR associate with
PCNA
(the proteins that keep the polymerases on DNA)
Mechanism of MMR in humans
(1)
error incorporated into newly synethizes strand
=> MSH2 and MSH6 bind to daughter strand (these proteins detect the mistake)
(2)
=> triggers binding and activity of MLH1 endocunclease (dimerized with PMS2), which make cuts near the mismatch.
MLH1 starts from inside (why “endo”) to make cut near the mismatch to mark it
the cuts flank the mutaiton
Then, DNA helicase unwinds and DNA exonuclease digests segment of the daughter strand
this helicase is different from the one used during DNA replication
(3)
Gap repair by Pol delta and DNA ligase
aka a single strand gap
nb. Pol delta is the lagging strand polymerase
What spontaneously occurs forming cytosine into uracil?
deamination
amine group is removed by hydrolysis (reaction with water) and the NH2 amine group is replaced with an oxygen
implication: C pairs with G wherease U pairs with A
error needs to be caught in time in order to not lead to bad pairing
Base Excision Repair (BER)
repairs small non-helix distorting lesions such as those from oxidation, deamination and alkylation
Mechanism of Base Excision Repair (BER)
(1)
DNA glycolase
recognizes the innapropriate or damaged DNA
hydrolyzes the bond between the mispaired base and the sugar phosphate backbone
after the removal of the base there is a gap
(2)
Apurinic/apyrimidinic endonuclease 1 (APE1) cuts the DNA backbone
at the site where ther is no base
(3)
AP lyase associated with DNA polymerase beta removes the deoxyribose phosphate
DNA pol beta is specific for repair
(4)
DNA polymerase beta fills the gap by acting like a polymerase and DNA ligase seals the nick in the sugar-phosphate backbone
nb. this system relies on double stranded DNA
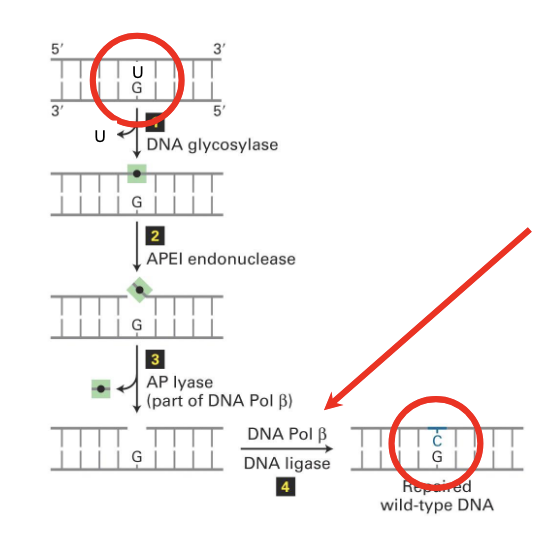
DNA glycolase
enzyme that performs the first step in BER by recognizing the and removing the damaged or inappropriate DNA bases from the DNA molecule
“glycolase” beause breaks the covalent bond of the base to the ribose (which is a sugar)
It provides specificity in repair — only removes the bases that need to be removed
there are different types of DNA glycolases in our cells
the one that removes uracil is called UNG
Bulky Lesions
modifications so big that they can change the regular double-stranded structure of DNA
often caused by photon of UV light
These lesions disrupt the normal double helix structure, blocking DNA replication and transcription
Often repaired by a system called nucleotide excision repair (NER)
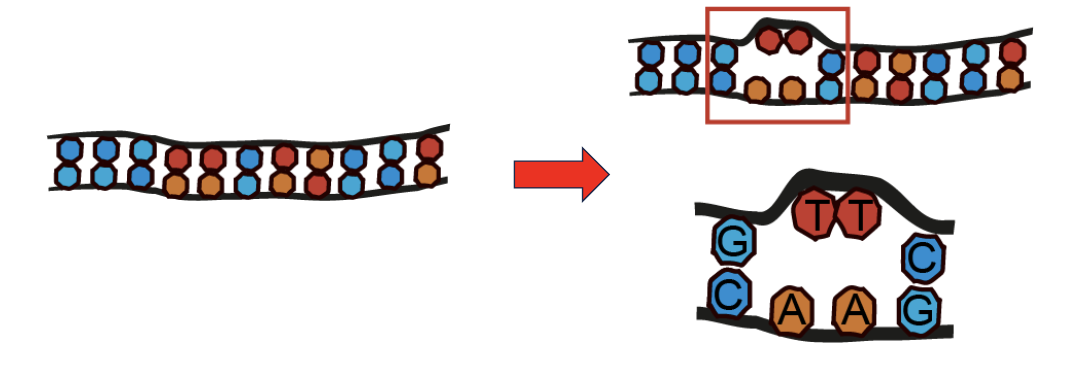
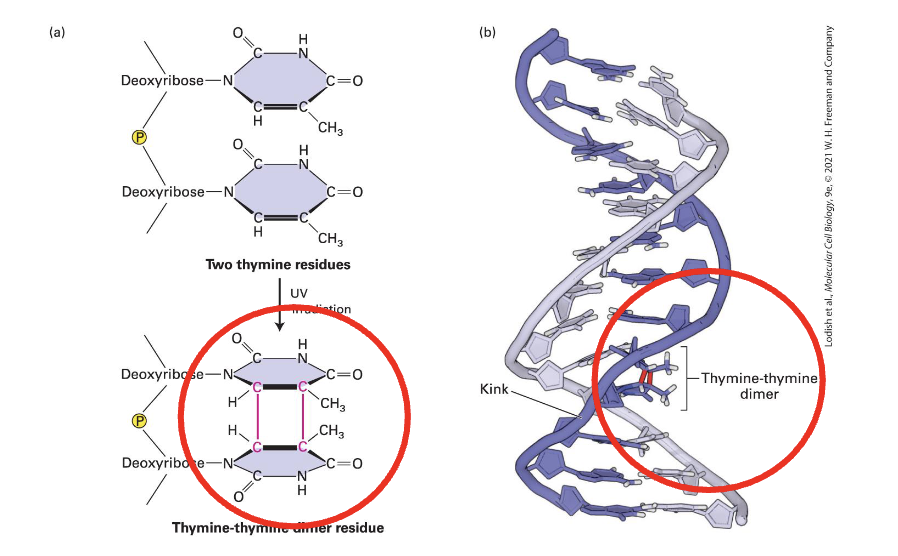
example of a bulky lesion: Thymine-thymine dimer (aka a pyrimidine dimer)
the two covalently linked thymines T-T form a rigid shape that distorts the DNA helix
like other bulky lesions, disrupts DNA replication and transcription
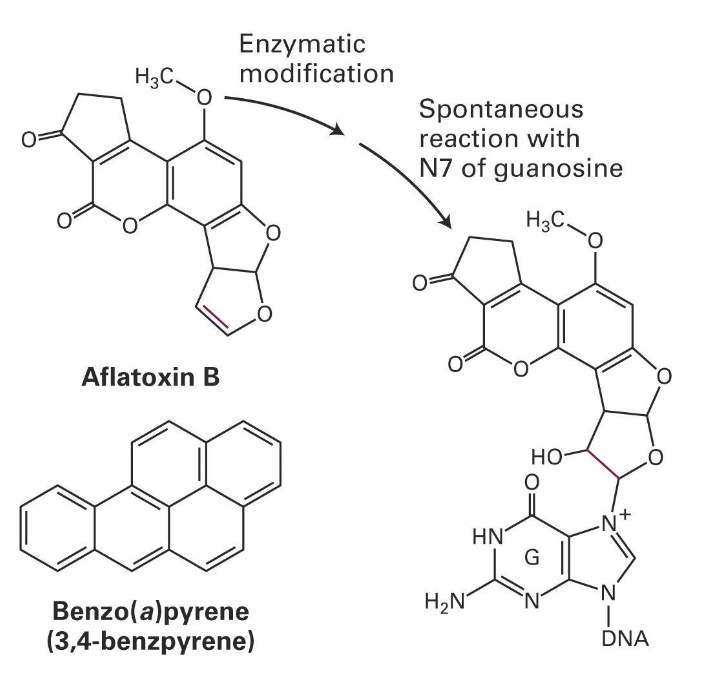
Bulky lesion example: from food contaminated by fungi Aspergillus or smoke from organic material (like cigarette or charboiled food)
Ingestion of Aflatoxin B
reacts with DNA and forms a covalent bond
enzymatic modification
spontaneous reaction with N7 of guanosine
benzo(a)pyrene (from the smoke of organic material) will generate modification similarly to Aspergillus to DNA
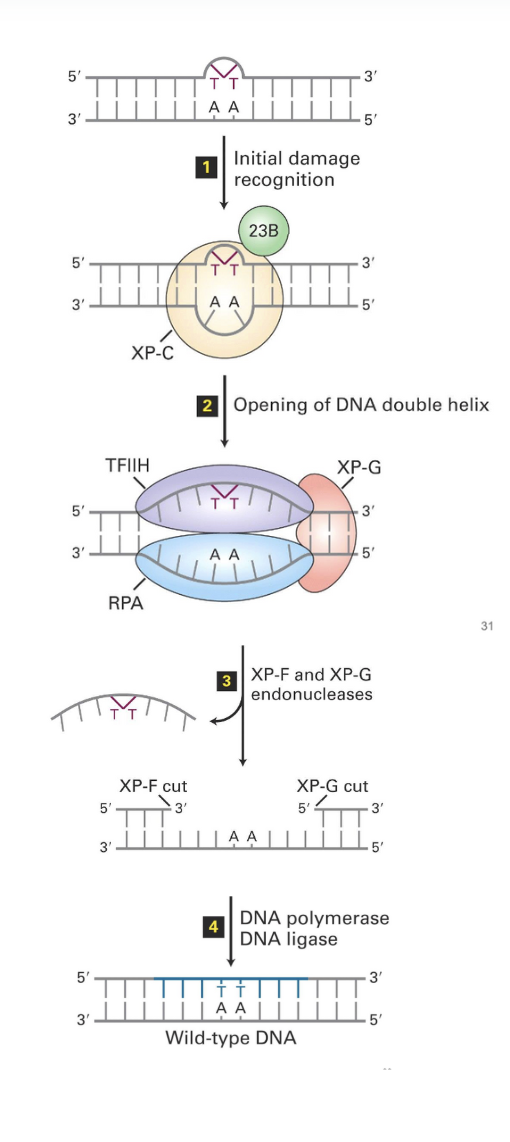
Nucleotide Excision Repair (NER) mechanism (for bulky lesions)
(1) initial damage recognition
A complex formed by proteins 23B and XP-C do the initial recognition of the DNA lesions
(2) opening of the DNA double helix
The complex of 23B and XP-C recruits the transcription factor TFIIH
TFIIH catalyzes the unwinding of the DNA region around the lesion
This recruits RPA and XP-G (which also have unwinding activity)
(nb. these are also used in DNA replication — pattern that proteins being used for replication also used for repair )
(3) XP-F and XP-G endonucleases
XP-F recruited
both XP-F and XP-G have endonuclease activities (aka they cut DNA)
generate cuts 24-32bp apart — a bigger gap is generated
releasing fragment carrying the lesion which will be degraded
(4) DNA polymerase & DNA ligase
the ssDNA gap generated is filled by DNA polymerase (probably Pol delta)
then joined to generate continuous strand by DNA ligase
nb. this system relies on double stranded DNA
What are example of humans Hereditary diseases and cancers associated with DNA Repair Defects
Hereditary nonpolyposis colorectal cancer
DNA mismatch repair affected
Xeroderma Pgmentosum
NER
what happens is a bulky lesion or other type of lesion is not repaired in time and the DNA is replicated
BER and NER repair mechanisms rely on double stranded DNA
if not caught in time the DNA polymerases will stop (stalled by replisome)
when replisome encounters DNA lesions that weren’t previously repaired the DNA polymerase of the replisome stops
this prevents the completion of DNA replication
big issue for cell because it dies
what repair mechanism is used as a last resort
Translesion Synthesis is a last resort solution for when replication
special type of DNA polymerase used: translesion(TLS) polymerases
what is special about translesion (TLS) polymerases
they are able to bypass the lesion using the damaged DNA as a template
however, often use the incorrect based at position opposite the lesion (because they have a single strand they don’t know the opposite base)
do not have proofreading exonuclease activity
hence they are error prone
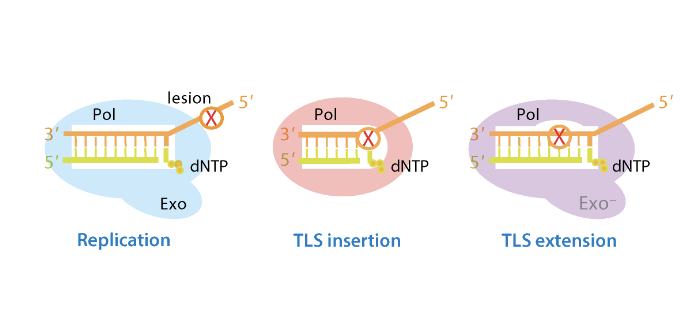
what is the implication of using TLS polymerases as a last resort (trade-off)
negative
easily make mistakes
leads to mutations
often at a great cost in humans
positive
mutations can be halful in bacteria