Bacteriology; Baileys and Scott (PARTIII_Sec.01: Chpt_12): Overview of Bacterial Identification Methods and Strategies
1/193
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
194 Terms
Microscopic identification of bacteria and its clinical specimen, Macroscopic identification of colony and its morphology, Additional microscopy for organism category identification
These are the steps in the entire workup of bacteria identification
Microscopic identification of bacteria and its clinical specimen
This step in the bacteria workup process involves providing the clinician with immediate therapy for life-threatening infections before further organism identification
Macroscopic identification of colony and its morphology
This step in the bacteria workup process involves identifying the type of hemolysis, pigment, size, texture, adherence to agar, pitting in agar, and other characteristics
Additional microscopy or gram stain observation
This step in the bacteria workup process involves the validation of the organism species and separating it into identifiable specie categories
Gram stain reaction, Cellular morphology or gram-positive or gram-negative bacteria
These are the bases for the categorization of organisms into cited broad categories
Catalase test
This is the test used to identify cellular morphology on gram-positive organisms
Oxidase test
This is the test used to identify cellular morphology on gram-negative organisms
MacConkey agar
This is the medium used for identified gram-negative rod or coccobacillus organisms
Catalase test, Oxidase test, MacConkey agar
These are helpful to the microbiologist in assigning the organism under observation to on of the primary cited categories
Identification of organisms based on common phenotypic traits shared with known members of the same genus or family
This is the clinical principle why microbiologist seem to be playing with the odds every day by finding the best biochemical fit and assigning the most probable organism identification
Neisseria species mixed testing results
This is the clinical rationale for the limitations of solely depending on flow charts for the identification process of organisms
HACEK organisms
This is the consideration of a microbiologist from an observed small-gram-negative rod in a Gram stain coming from a blood specimen with a patient having an endocarditis
Haemophilus spp., Aggregatibacter spp. Cardiobacterium spp., Eikenella corrodens, Kingella spp.
These are a group of gram-negative bacilli commonly known as the HACEK organisms
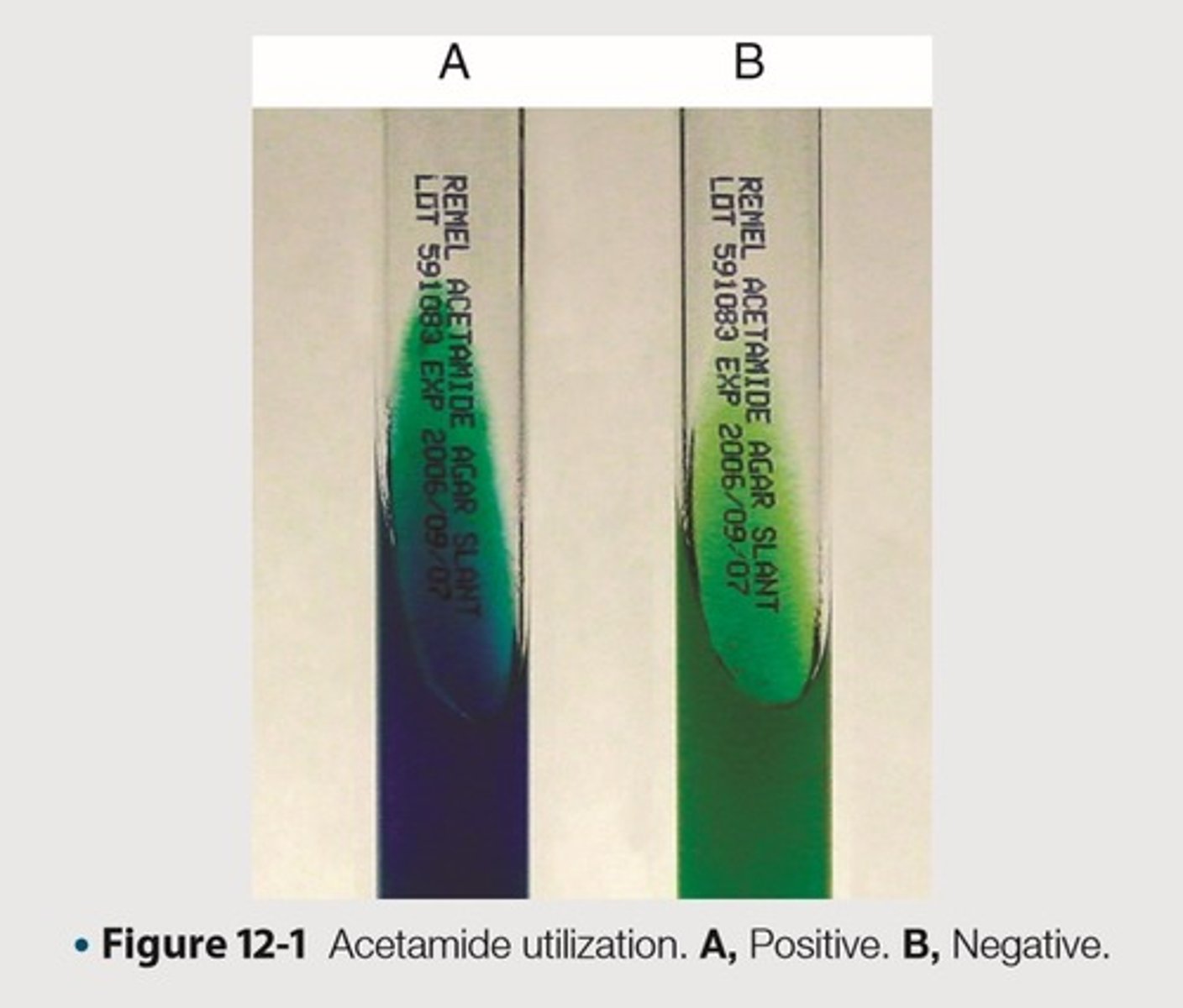
Differentiate microorganisms based on the ability to use acetamide as the sole source of carbon
This is the purpose of Acetamide utilization
Bacteria capable of growth on this medium produce the enzyme acylamidase
This is the principle of Actetamide utilization
5g of sodium chloride, 1g of ammonium dihydrogen phosphate, 1g of Dipotassium phosphate, 15g of agar, 0.8g of bromothymol blue indicator per 1000 mL, 10g of acetamide, 6.8 pH
This is the media contents of the agar or broth used in Actetamide utilization
Deaminates acetamide to release ammonia
This is the clinical significance of bacteria capable of growing on an Actetamide utilization medium to produce the enzyme acylamidase
Production of ammonia results in an alkaline pH
This is the clinical significance of bacteria producing acylamidase to deaminate acetamide to release ammonia
Causes the medium to change color from green to royal blue
This is the clinical significance of bacteria producing acylamidase to deaminate acetamide to release ammonia causing an alkaline power of hydrogen
Inoculate acetamide slant with a needle using growth from an 18- to 24-hour culture
This is the first methodical step for Acetamide utilization
Bacterial growth will be too heavy
This is the methodological significance of not inoculating from a broth culture instead of an 18-24 hour culture in Acetamide utilization
Incubate aerobically at 35-37 degrees Celsius for up to 4 days
This is the second methodical step for Acetamide utilization
Equivocal
This pertains to test results that could not be interpreted as Positive or Negative
Equivocal Acetamide slant
This is the methodological significance of reincubating an Acetamide slant for an additional 2 days
Reincubate for an additional 2 days
This is the number of days in reincubating an equivocal Acetamide slant
Blue color
This is the expected color result for a Positive Acetamide utilization
No color change
This is the expected color result for a Negative Acetamide utilization
Deamination of the acetamide
This is the methodological rationale for a blue colored result in Acetamide utilization
Bacterial growth without a color change may indicate a positive test result
This is the limitation of Acetamide utilization in bacterial growth results (?)
If further incubation results in no color change, repeat test with less inoculum
This is the limitation of Acetamide utilization in methodological incubation of inoculum (?)
Inoculum
This is the introduction of microbes into medium
Pseudomonas aeruginosa-growth, blue color
This is the organism and its expected result for the quality control of acetamide utilization to exhibit positive results
Escherichia coli-no growth, green color
This is the organism and its expected results for the quality control of acetamide utilization to exhibit negative results
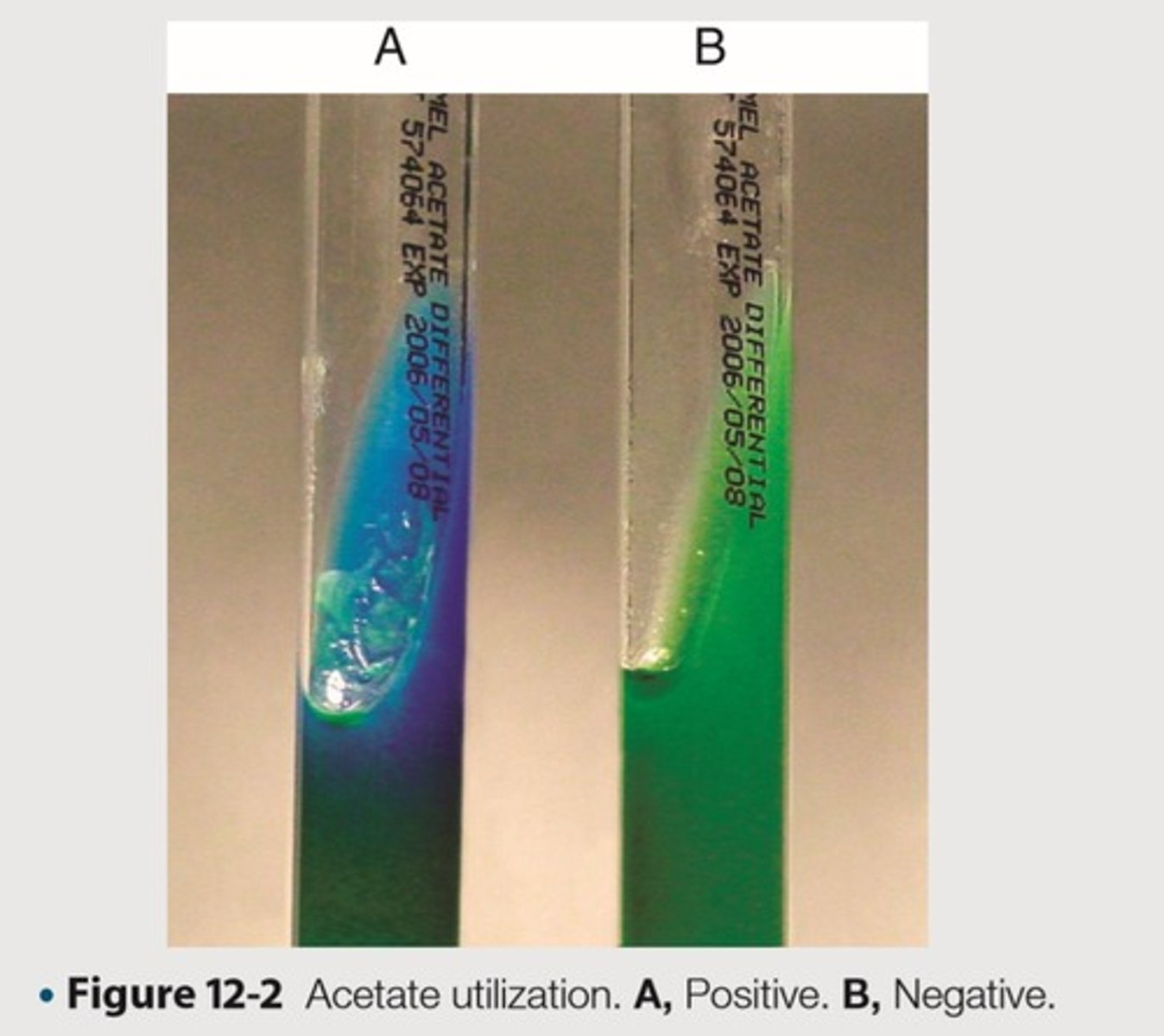
Differentiate organisms based on ability to use acetate as the sole source of carbon
This is the purpose of Acetate utilization
Generally used to differentiate Shigella spp. and Escherichia coli
This is the general purpose of Acetate utilization
Organisms capable of using sodium acetate to grow on prepared medium
This is the clinical principle of Acetate utilization
Resulting in an alkaline pH
This is the clinical significance of organisms using sodium acetate for growth (?)
Turns the indicator from green to blue
This is the clinical significance of a Acetate utilization medium resulting to an alkaline pH
2g of Sodium acetate (NaC2H3O2), 0.1g of Magnesium sulfate (MgSO4), 1g of Ammonium dihydrogen phosphate, 20g of agar, 0.8g of bromothymol blue indicator per 1000 mL, pH 6.7
This is the media contents of the agar or broth used in Acetate utilization
Inoculate slant lightly with a straight inoculating needle from an 18- to 24-hour culture
This is the first methodological step of Acetate utilization
Bacterial growth will be too heavy
This is the methodological significance of not inoculating from a broth culture instead of an 18-24 hour culture in Acetate utilization
Incubate at 35-37 degrees Celsius for up to 7 days
This is the second methodological step of Acetate utilization
Medium becomes alkalinized, blue color
This is the expected positive result in Acetate utilization
No growth or growth with no indicator change to blue
This is the expected negative result in Acetate utilization
Some strains of E. coli may use acetate at a very slow rate or not at all
This is the limitation of Acetate utilization
Resulting in a false negative in the identification process
This is the clinical rationale why Acetate utilization could be limited because of E. coli
Escherichia coli-growth, blue color
This is the organism and its expected results for the quality control of Acetate utilization to exhibit positive results
Shigella sonnei-small amount of growth, green color
This is the organism and its expected results for the quality control of Acetate utilization to exhibit negative results
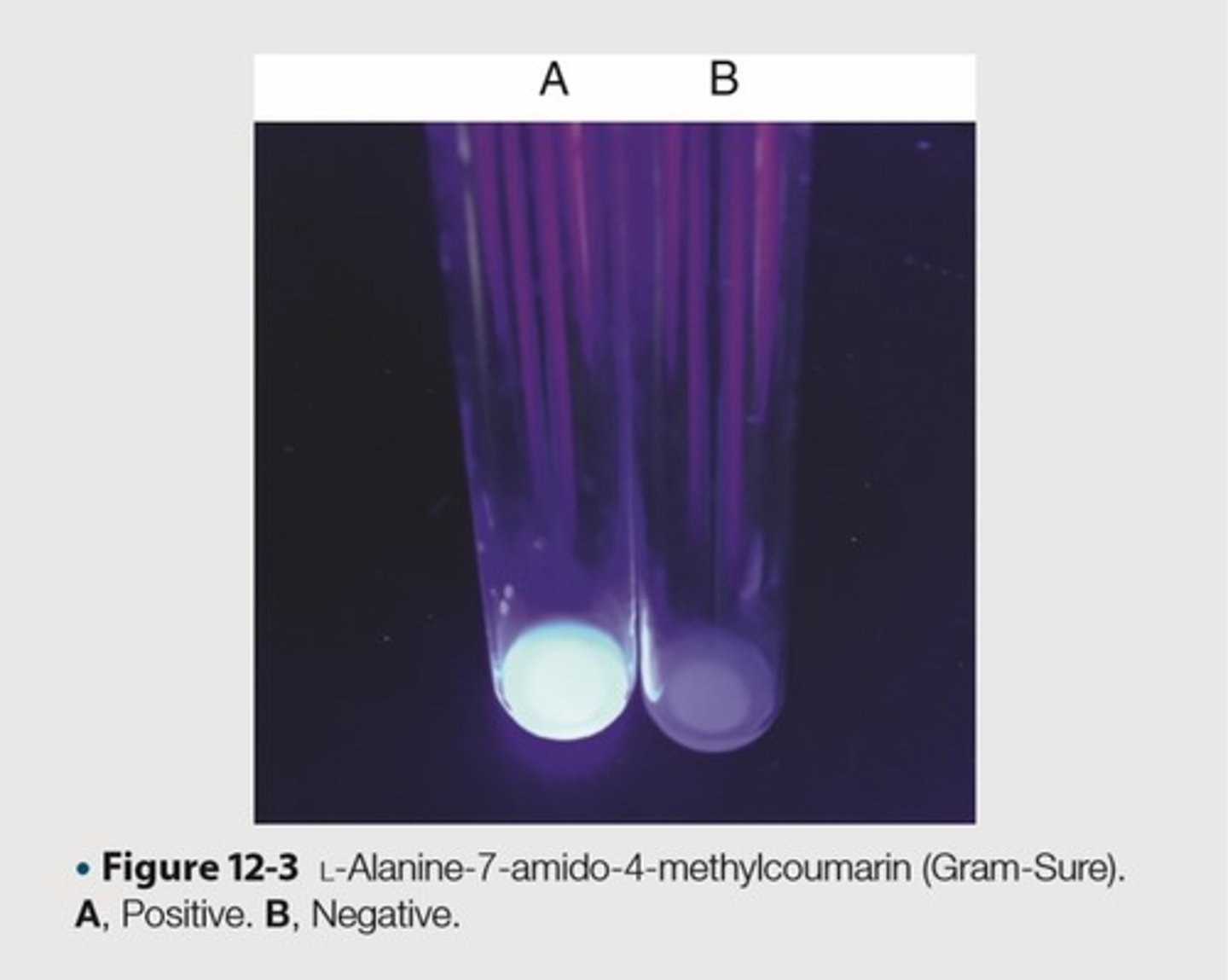
Test used in conjunction with Gram stain to distinguish aerobic gram-positive rods or coccobacilli
This is the test purpose of L-Alanine-7-amino-4-methylcourmarin (Gram-Sure)
Aerobic gram-positive rods or coccobacilli may appear gram-negative or gram-variable
This is the clinical rationale for distinguishing aerobic gram-positive rods or coccobacilli in L-Alanine-7-amino-4-methylcourmarin (Gram-Sure)
L-Alanine-7-amino-4-methylcourmarin
This is the compound impregnated in a commercially prepared disk, Remel-Thermo Fisher Scientific in Gram Sure
Remel-Thermo Fisher Scientific, Lenexa, KS
This is the commercially prepared disk impregnated with L-Alanine-7-amino-4-methylcourmarin and hydrolyzed by aminopeptidase
Gram-negative organisms produce an aminopeptidase
This is the principle used in L-Alanine-7-amino-4-methylcourmarin (Gram Sure)
Capable of hydrolyzing the reagent in the disk
This is the clinical rationale of aminopeptidase in L-Alanine-7-amino-4-methylcourmarin (Gram Sure)
Forming a blue fluorescent compound that is visible under long-wave UV light
This is the clinical rationale of hydrolyzing L-Alanine-7-amino-4-methylcourmarin with aminopeptidase in Remel-Thermo Fisher Scientific, commercial disk
Inoculate a pure colony of overnight growth to 0.25 mL of demineralized water in a clean 12 by 72 mm test tube
This is the first methodological step in L-Alanine-7-amino-4-methylcourmarin (Gram Sure)
16-18 hours after initial culture
This is the duration of the overnight growth of the organism in L-Alanine-7-amino-4-methylcourmarin (Gram Sure)
Place a Gram-Sure disk in the emulsion
This is the second methodological step in L-Alanine-7-amino-4-methylcourmarin (Gram Sure)
Incubate emulsifying Gram-Sure disk at room temperature for 5-10 minutes
This is the third methodological step in L-Alanine-7-amino-4-methylcourmarin (Gram Sure)
Observe blue fluorescence by placing the tube under long-wave UV light
This is the fourth methodological step in L-Alanine-7-amino-4-methylcourmarin (Gram Sure)
Fluorescent or blue
This is the expected result for aerobic, gram-negative rods and coccobacilli
Colorless
This is the expected result for aerobic, gram-positive rods and coccobacilli
Obligate anaerobic organisms may fail to give expected results
This is the limitation in L-Alanine-7-amino-4-methylcourmarin (Gram Sure)
Escherichia coli-blue fluorescence
This is the organism and its expected results for the quality control of L-Alanine-7-amino-4-methylcourmarin (Gram Sure) to exhibit positive results
Staphylococcus aureus-no fluorescence
This is the organism and its expected results for the quality control of L-Alanine-7-amino-4-methylcourmarin (Gram Sure) to exhibit negative results
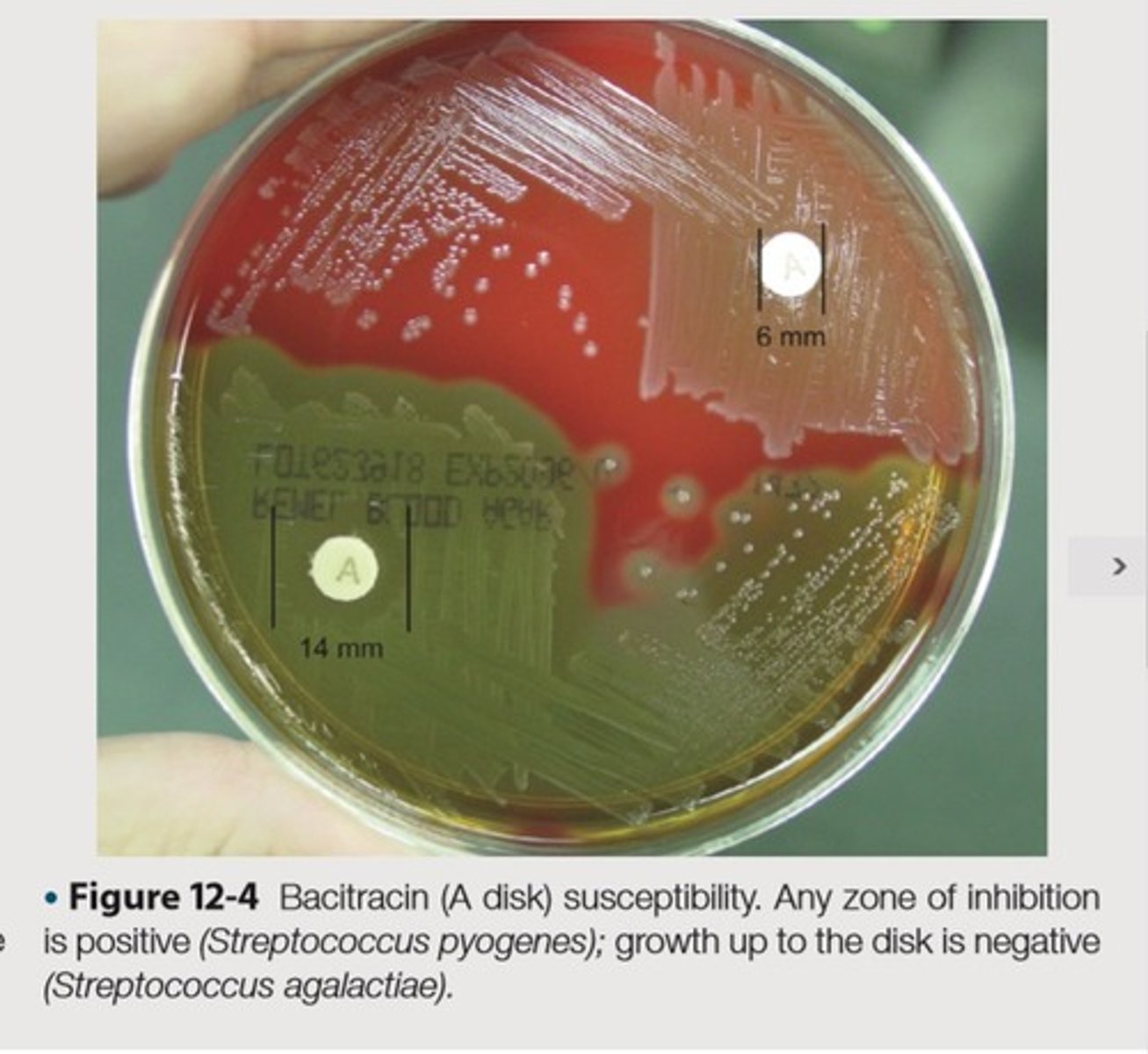
Presumptive identification and differentiation of beta-hemolytic group A streptococci from other beta-hemolytic streptococci, Distinguish staphylococci species from micrococci
This is the purpose(2) of Bacitracin Susceptibility
The antibiotic bacitracin inhibits the synthesis of bacterial cell walls
This is the principle of Bacitracin Susceptibility
0.04 units of bacitracin is impregnated into TaxoA disk and placed on an agar plate
This is the methdological principle of the utility of bacitracin in Bacitracin Susceptibility
Allowing the antibiotic to diffuse into the medium and inhibit the growth of susceptible organisms
This is the methodological rationale of impregnating bacitracin into TaxoA disk and placed on an agar plate
Examined for zones of inhibition surrounding the disks
This is the clinical observation to be made after incubation on the inoculated plates of Bacitracin Susceptibility
Using an inoculating loop, streak two or three suspect colonies of a pure culture onto a blood agar plate
This is the first methodological step of Bacitracin Susceptibility
Using heated forceps, place a bacitracin disk in the first quadrant, gently tap the disk to ensure adequate contact with agar surface
This is the second methodological step of Bacitracin Susceptibility
First quadrant
This is the area of heaviest growth in the blood agar plate of Bacitracin Susceptibility
Look for a zone of inhibition around the disk
This is the fourth methodological step of Bacitracin Susceptibility
Incubate the plate for 18-24 hours at 35-37 degrees Celsius in ambient air for staphylococci and in 5-10% CO2
This is the third methodological step of Bacitracin Susceptibility
Streptococci differentiation
This is the methodological rationale of third methodological step of Bacitracin Susceptibility
Any zone of inhibition greater than 10 mm; susceptible
No zone of inhibition; resistant
Performance depends on the integrity of the disk
These are the limitations of Bacitracin Susceptibility
Proper storage and expiration dates should be maintained
This is the resolution for the limitation of Bacitracin Susceptibility
Streptococcus pyogenes-susceptible, Micrococcus luteus-susceptible
This is the organism and its expected results for the quality control of Bacitracin Susceptibility to exhibit positive results
Streptococcus agalactiae-resistant, Staphylococcus aureus-resistant
This is the organism and its expected results for the quality control of Bacitracin Susceptibility to exhibit negative results
Presumptive identification of enterococci and organisms in the Streptococcus bovis group; Differentiates enterococci and group D streptococci from non-group D viridands streptococci
This is the purpose(2) of Bile Esculin Test
Inhibition of Gram-positive bacteria, Growth of organisms capable to hydrolyze esculin to esculetin
This is the principle of Bile Esculin Test
4% bile
This is the percentage composition of bile used in Bile Esculin Test
Reacts with Fe3+ and forms a dark brown to black precipitate
This is the clinical significance of esculetin in Bile Esculin Test
11g of beef extract, 34.5g of enzymatic digest of gelatin, 1g of esculin, 2g of ox bile, 0.5g of ferric amonium citrate, 15g of agar per 1000 mL, pH 6.6
This is the medium composition of the agar slant used in Bile Esculin Test
Inoculate one to two colonies from an 18-24 hour culture onto the surface of the slant
This is the first methodological step in Bile Esculin Test
Incubate at 35-37 degrees Celsius in ambient air for 48 hours
This is the second methodological step in Bile Esculin Test
Growth and blackening of the agar slant
This is the expected positive result of Bile Esculin Test
Growth and no blackening of medium (not shown)
This is the expected negative result of Bile Esculin Test
Some organisms may grow poorly or not at all on this medium, because of nutritional requirements
This is the limitation of Bile Esculin Test
Enterococcus faecalis-growth, black precipitate
This is the organism and its expected results for the quality control of Bile Esculin Test to exhibit positive results
Escherichia coli-growth, no color change; Streptococcus progenes-no growth, no color change
This is the organism and its expected results for the quality control of Bile Esculin Test to exhibit negative results

Differentiates Streptococcus pneumoniae from alpha-hemolytic streptococci
This is the purpose of Bile Solubility Test
Bile or a solution of a bile salt rapidly lyses pneumococcal colonies
This is the principle of Bile Solubility test
Sodium deoxycholate
This is an example of a bile salt used in the bile solubility test
Amidase
This is the intracellular autolytic enzyme wherein the presence of it causes the intensity of lysis of organism in Bile Solubility Test
Lowers the surface tension between the bacterial cell membrane and medium
This is the clinical rationale of bile salts
Accelerates the organism's natural autolytic process
This is the clinical rationale of lowering the surface tension between the bacterial cell membrane and medium