Lecture 4: DNA Packaging and Cell Division
1/53
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
54 Terms
What does DNA look like in prokaryotes
single circular DNA and non-chromosomal DNA/plasmids
gene
segment of DNA that codes for a protein or RNA product
sister chromatid
identical copies in replicated chromosome
replicated chromosome
a chromosome comprised of two identical DNA molecules, joined together
homologous chromosomes
different versions of same chromosome type in the same cell
maternal and paternal versions of each chromosome
chromatin
complex of DNA and protein
nucleosomes are further packed on top of one another to generate a more compact structure called chromatin fiber
human cells contain ___ copies of every chromosome
2
one from dad and one from mom
supercoiling
DNA more compact
occurs in eukaryotes and prokaryotes
negative supercoiling: opposite direction to helix
positive supercoiling: same direction as helix
supercoiling can occur spontaneously during DNA replication
supercoiling controlled by topoisomerase
temporarily breaks and rejoins DNA strands to relieve stress
Detection of supercoiling
DNA gel, put DNA in each lane, some of it is relaxed, cut (linear) or supercoiled
DNA travels at different speeds
slowest to fastest: relaxed, linear, supercoiled
supercoiled moves faster because more compact and denser
Compaction of the Bacterial Chromosome
relaxed DNA is circular in prokaryotes
DNA-binding proteins can bind to circular chromosome
DNA becomes supercoiled into loops held in place by DNA-binding proteins which stabilize structure
Eukaryote Nucleus
density is not uniform
euchromatin
lighter area
less packed/dense
interphase
heterochromatin
darker area
more packed/dense
mitosis
nucleolus
special area where ribosomes are made
level of compaction controls _____
transcription
genome size
number of base pairs
Prokaryotes genome
106 base pairs
one circular chromosome + non chromosome DNA (plasmids)
Eukaryotes genome
linear chromosomes + non-nuclear DNA (mitochondrial)
107 bp fungi
109 bp mammals
3 × 109 bp humans
species with similar genome sizes can have ____
very different chromosome numbers and sizes
interphase
when chromosomes are duplicated = DNA replication
cells express many genes (transcription and translation)
cells are long thin threads of DNA that can’t easily be distinguished
chromosomes are less condensed but still considered tightly packed
mitosis
duplicated chromosomes are highly condensed
DNA is compacted ≥ 15000 fold in mitotic human chromosomes
theorized that chromosomes condense so don’t get tangled during metaphase
gene expression ceases (transcription and translation)
nuclear envelope breaks down
Replication Origins
multiple origins in eukaryotes
one origin in prokaryotes
telomeres
repetitive nucleotide sequence that caps the ends of linear chromosomes
helps chromosomes not shorten with each round of replication
protective caps that keep chromosome from bring mistaken as broken DNA
two telomeres
centromeres
specialized DNA sequence that allows duplicated chromosomes to be separated during M phase
nucleosome
nucleosome consists of 8 histone proteins: two H2A, two H2B, two H3, and two H4
refers to both core particle and linker DNA
DNA wrapped around histone two times
~200 bp of DNA
beads on a string
first level of chromatin packing
histones
highly conserved proteins around which DNA wraps to form nucleosomes
why do histones stick to DNA
high proportion of positively charged amino acids help bind to negative phosphate on DNA
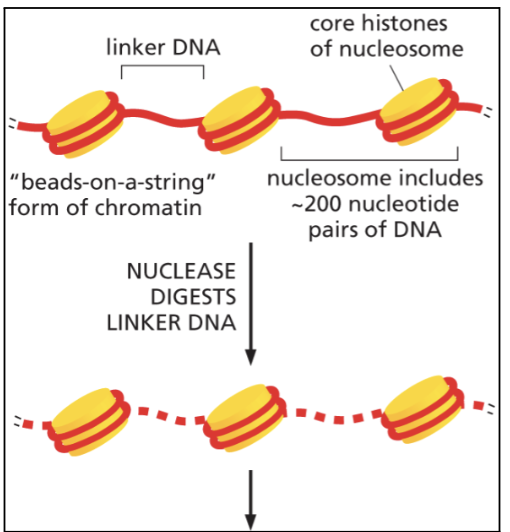
how many bp of DNA around core particle
146 bp
how many base pairs of DNA around linker DNA?
~ 44 bp
nucleosomes can slide together or apart so number changes
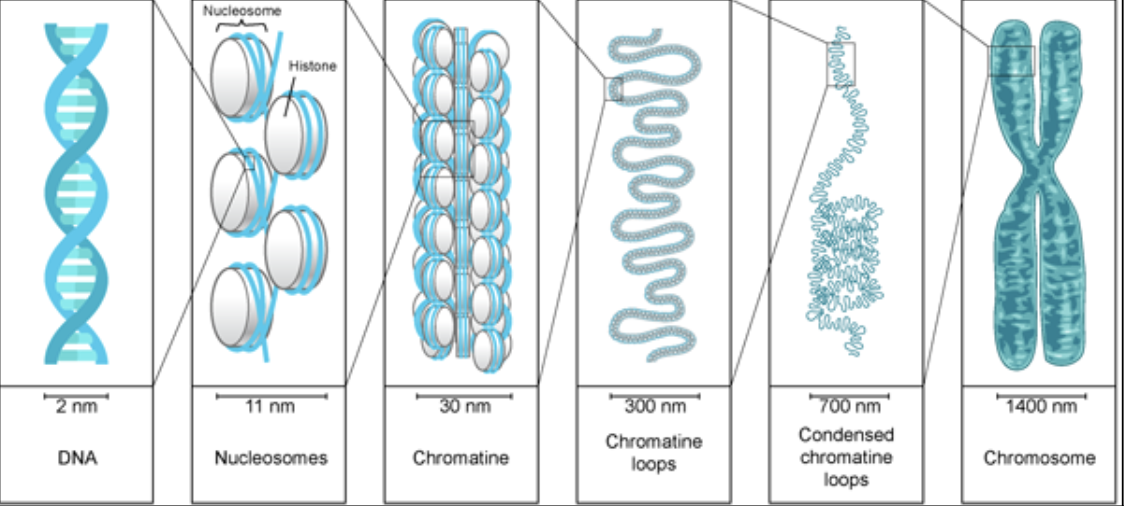
incorrect hierarchical model of Chromatin compaction
said compaction was a regular ordered process
in reality: disordered (not tangled) chromatin chains
different configurations and densities each time
width is thicker in mitotic chromosomes
Cell cycle
orderly sequence of events by which a cell duplicates and divides
G1 (Interphase)
1) cell grows
2) organelles replicated
3) high rates of transcription and translation
during transcription nucleosomes are broken apart to expose certain genes called “open” DNA
S (Interphase)
1) when chromosomes are duplicated
G2 (Interphase)
cell grows
do transcription and translation occur during all of interphase
yes
mitosis length
A briefer stage when duplicated chromosomes are distributed into two daughter nuclei
cell cycle control system
Makes sure each process is complete before next begins
Cell cycle checkpoints
1) Late G1
environment is favorable for proliferation
can enter G0 if not
2) End of G2
make sure DNA is fully replicated
3) Midway through Mitosis
make sure chromosomes properly attached to mitotic spindle
What does the cell-cycle control system depend on?
cyclically activated protein kinases (maturation-promoting factor)
phosphorylate and dephosphorylate proteins
turns proteins off and on
how does phosphorylation work?
phosphorylation done by kinases
phosphate attached to side chain of amino acids
dephosphorylation done by phosphatases
Cyclin
discovered by Tim hunt using SDS-Page on Sea Urchins
regulatory protein whose concentration rises and falls at specific times
helps control progression from one state of cell cycle to next by binding to cyclin dependent kinase
concentration varies in cyclical fashion
rise in concentration due to continued transcription of cyclin gene
decline in cyclin concentration due to targeted destruction of the protein
how does a cell progress through various stages of the cell cycle?
controlled expression and degradation of various cyclin proteins
is the same cyclin-cdk used for all steps in cell cycle
no, cyclin B-cdk complex promotes mitosis but there are others
Activated CDK phosphorylate different proteins that are responsible for…
1) nuclear membrane degradation
2) microtubule assembly
3) chromatin condensation
Do levels of CDK change?
no
centrosomes
are duplicated before mitosis
principal microtubule-organizing center
house the centrioles
when mitosis begins, two centrosomes separate
mitotic spindle
form from microtubules and other proteins during prophase
individual filaments alternate between growing and shrinking: dynamic instability
3 types of microtubules
1) kinetochore
reach out from centrosome to center of chromosome
multiple microtubules can attach here
2) aster
spread out backwards from centrosome and will reattach to nuclear membrane when it reforms
3) polar
don’t attach to chromosomes but to each other
kinetochore
a protein complex associated with centromere
each duplicated chromosome has two kinetochores: one on each sister chromatid
Prophase (1st step of Mitosis)
chromosomes condense
mitotic spindle assembles outside nucleus between two centrosomes which have begun to move apart
Prometaphase (2nd step of Mitosis)
breakdown of nuclear envelope
chromosomes attach to spindle microtubules via kinetochore
motor proteins composed of kinesin or dynesin pull microtubules and chromosomes in position
Metaphase (3rd step of Mitosis)
chromosomes align in equator
Anaphase (4th step of Mitosis)
Sister chromatids separate and are pulled toward spindle pole
Pushing and pulling force
Push spinde poles from each other and pull spindle poles towards reforming cell membrane
Telophase (5th step of Mitosis)
two sets of chromosomes arrive at poles of spindle
new nuclear membrane reassembles around each set
Cytokinesis (6th step of Mitosis)
cell divides
microtubules
extend through cytoplasm
can rapidly assemble and dissassemble
form cilia or flagella
microtubule structure
polymers whose building blocks are tubulin alpha and beta dimers
hollow and spiral tube
+ and - negative orientation ends
- end anchored to centrosome
where you subtract alphabeta subunits
+ end attaches to centromere
where you add alphabeta subunits
organize in centrosome