Week 8: Evolution of Gene Regulation + Chromosome Evolution
1/55
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
56 Terms
How to measure changes in gene regulation?
Use RNA sequencing
Isolate mRNA from tissue of interest
Sequence all mRNA in a sample
Count number of sequences mapping to each gene =expression level
Interpreting gene expression data
Data is displayed in a grid where each row represents a gene and each column represents a sample taken from a different tissue, stage, species or environmental condition
Red represents up-regulated genes and green represents down-regulated genes. Black represents unchanged expression.
The heatmap may also be combined with clustering methods which group genes together based on the similarity of their gene expression pattern.

Highly conserved gene regulation
specific sequences + regulatory mechanisms involved in controlling gene expression remain relatively unchanged across different species/ within a single organism over evolutionary time
What can gene regulation allow organisms to do?
Respond to changing environments
Links to phenotypic plasticity
Example: Killifish gene expression responds to ecological variation
Gene regulation and adaptive evolution
Beak size in Darwin’s Finches varies among species and is key to ecological niches.
Crushing beaks → wider/ larger than probing beaks
Whitehead et al. 2011 PNAS
Experiment looked at differences in expression in stages of beak development + compared crushing + probing beak
How can beak shape vary so much in species with very little genetic difference?
Bmp4 expressed in crushing beak early
Is all gene expression adaptive?
No
Across 5,636 genes divergence from human is roughly clock-like (testis genes are an exception) →Suggests most change in gene expression due to drift
Genes not under selection → change at same rate
Brawand et al. 2011
Signatures of lineage-specific adaptive change
Romero et al. 2012 Nature Reviews Genetics
Studies use RNAseq to compare the expression of orthologous genes in different groups of mammals RNA can be extracted from various tissues (e.g prefrontal cortex cerebellum, heart liver and testes) and relative expression levels measured.
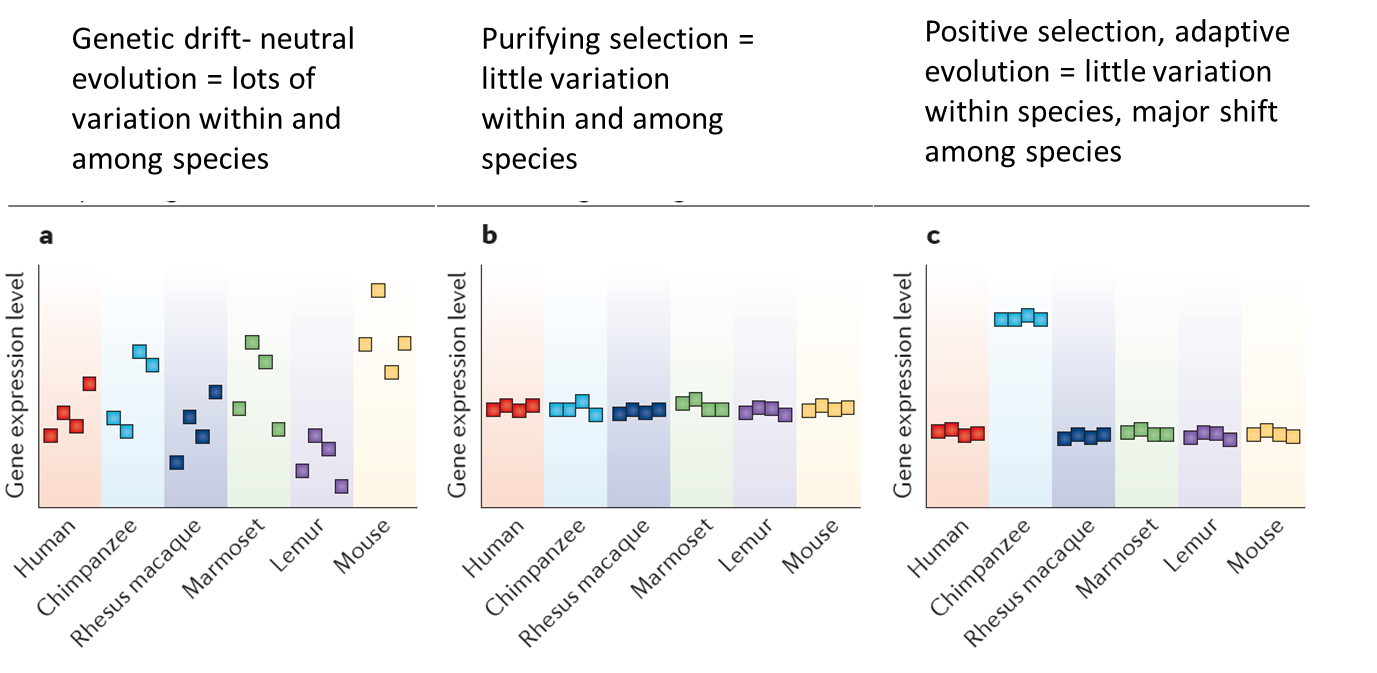
Human-specific adaptive expression shifts
Modules with specific expression states in human brain (prefrontal cortex; 259 genes) and primate cerebellum (189 genes) are shown.
Bars represent the weighted average expression of all genes in a module, for each sample (horizontal grey line indicates average bar height).
Samples above the red line are considered to have a distinct expression state.
The large number of gene ontology categories related to neural ensheathment etc probably reflects the larger proportion of myelinated axons (white matter) in the human prefrontal cortex than in that of other primates
Pre-Frontal cortex important for primates
Brawand et al. 2011 Nature
graph = number of gene pathways/ modules with lineage-specific adaptive shifts
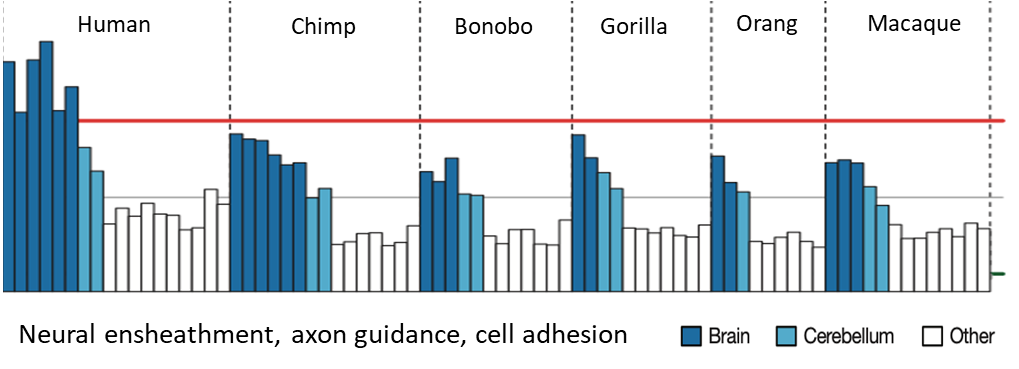
Human-specific adaptive expression shifts at cellular resolution (more recent study)
Caglayan et al. (2023) Nature
Comparative studies of whole brain region expression have identified genes/pathways that affect synaptogneisis and myelination
Brodmann area 23 is part of the posterior cingulate cortex, involved in higher-order cognitive processes such as theory of mind. Single cell RNAseq analysis comparing cells from this region in Humans, Chimpanzees and Macaques carried out
Results suggest that an evolutionary modification in human brain may have been achieved through loss-of-expression mutations of genes (e.g. SH3RF3) expressed in oligodendrocyte progenitors that delay their maturation and prolong brain plasticity
Most supported theory of sex chromosome evolution
Many genes under “sex-antagonistic selection.” Different alleles are favoured in females and males.
Genetic factors controlling the male coloration traits in guppies are concentrated on the male-determining factor carrying chromosome
e.g males → colourful → conspicuous to predators but more conspicuous to mates → outweighs negative
females → does not outweigh
Selection favours decreased recombination between genes under sex-antagonistic selection and the locus that determines sex where this keeps male-favouring alleles in cis with the male-determining factor (and vice versa for females)
Inversion mutation → suppress recombination
^ long term deleterious side effects
Invert region around sex determining factor → colouring genes can’t recombine onto female chromosome
What leads to Y chromosome decay?
Reduced recombination between proto-X and Y
Muller’s Ratchet
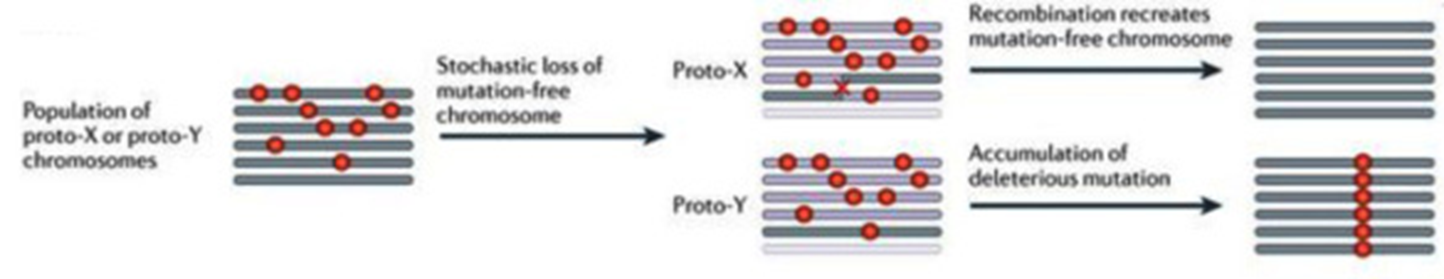
Genetic Hitchhiking
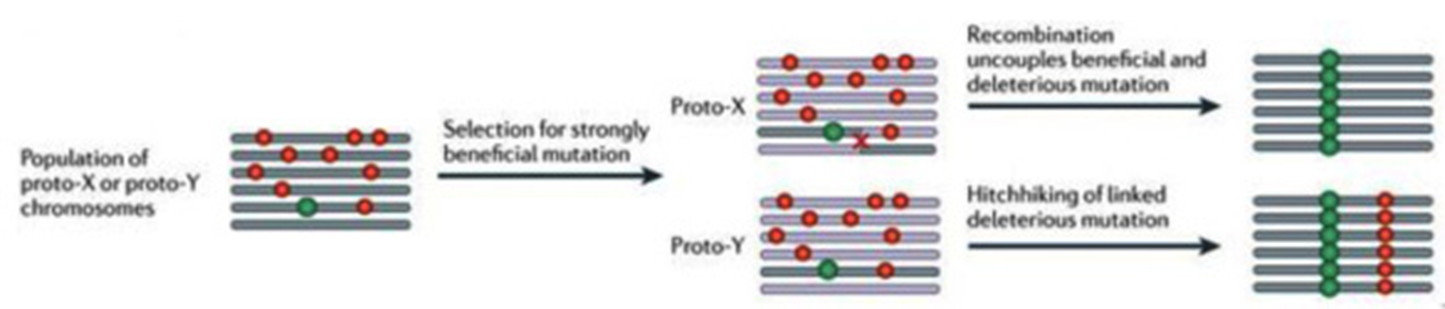
Sex chromosome evolution
Major and minor sex chromosome pairs start as homologous autosomes
Origin of new sex determining gene on one chromosome
When near a sexually antagonistic locus selection for inversions which prevents recombination
Without recombination, minor orthologs loses non-essential genes
New regions of the Y brought in proximity to sex determining gene
Repeat across length of chromosomes
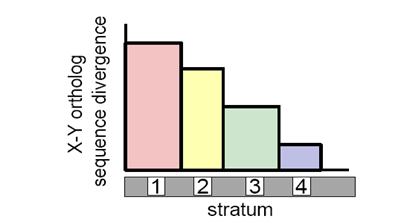
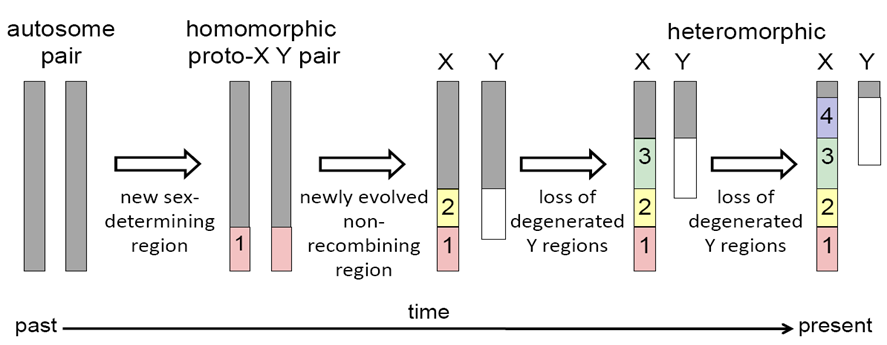
Exceptions to the classical model of sex chromosome evolution
Reduction of Y chromosome true in mammals → species of shrew → lost Y chromosome but produces males through different form of sex determination
In many non-model organisms new methods for detecting sex-linked sequences indicate that ancestral sex chromosomes have reverted to autosomes, and been replaced by a new set of sex-determining chromosomes (sex-chromosome turnover)
Clades where recurrent non-homologous sex-chromosome turnover has been detected
African cichlids
True frogs
Flies
Medaka fishes
African clawed frogs
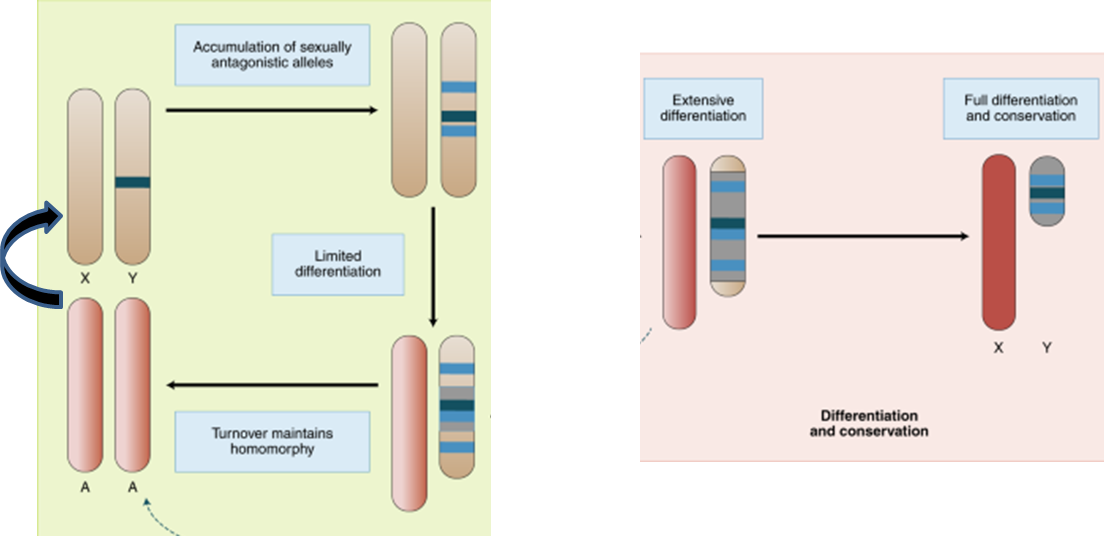
Modelling sex-chromosome turnover and conservation
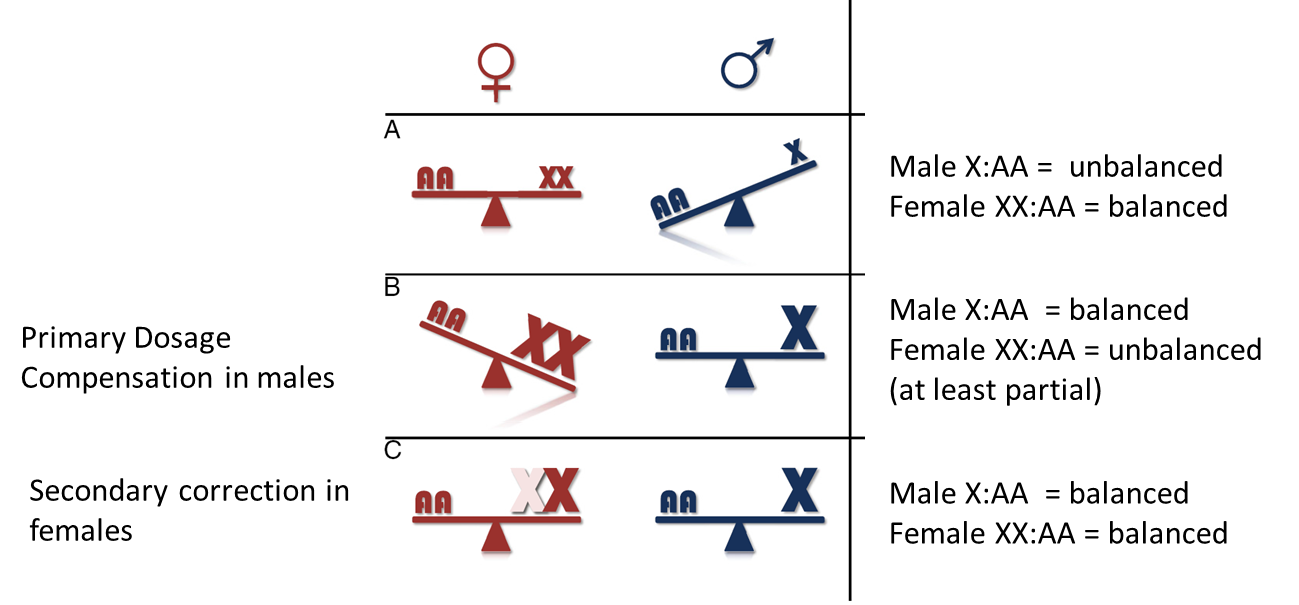
Why is male: female X chromosome dosage compensation needed?
Sex chromosome divergence means X chromosome copy number is different in males and females
Copy number correlates to transcription and translation rate
Loss of Y genes leaves males with just one functional X chromosome = ½ original gene dose
Level of gene expression = function of the number of copies of genes
Y chromosome shrinking → males have 1 copy → half the dose
Potentially large phenotypic consequences to the heterogametic sex
How are sex chromosomes regulated?
Dosage Compensation
Hyper-transcription of male X
Hypo-transcription of both X’s in females
X chromosome inactivation in females
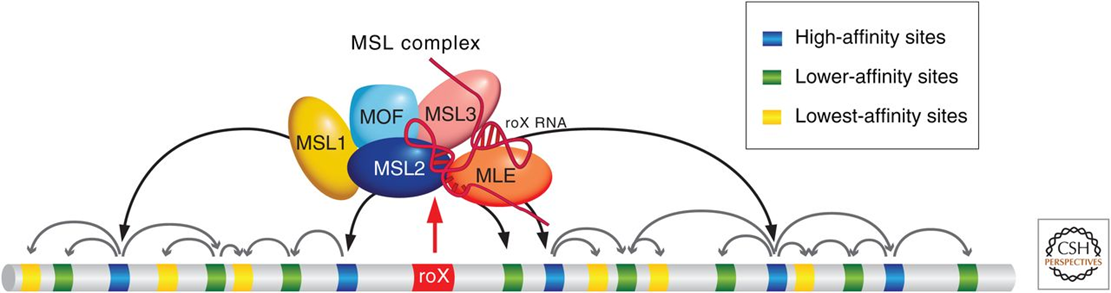
Dosage compensation in Drosophila
Lucches and Kuroda (2015) CSH Perspect. Biol.
Hyper-transcription of male X
Xmale= Xfemale
Xmale=Automale
MSL complex binds to regions containing transcriptionally active promoter and chromatin modifications (marks) mediate increased transcription at those promoters
How is MSL binding/activity targeted to a single chromosome? Involves roX non-coding RNAs and in-cis spreading from specific initiation sites
Sex determination in Drosophila
different mechanism for sex determination
females = XX
males = X + tiny Y with hardly any genes
Dosage Compensation in C. elegans
Lau & Csankovszki (2015). Curr. Opin. Genet. Dev
2,801 genes on X
20,176 genes total
= 14% of functional genome
no Y chromosome
Hyper-transcription of X in males & hermaphrodites by unknown mechanism
Xmale=Automale
Xmale= ½ Xherm
Xherm=2Autoherm
DCC mediated hypo-transcription of X in hermaphrodites
Xfemale=Autoherm
Xmale=Xherm
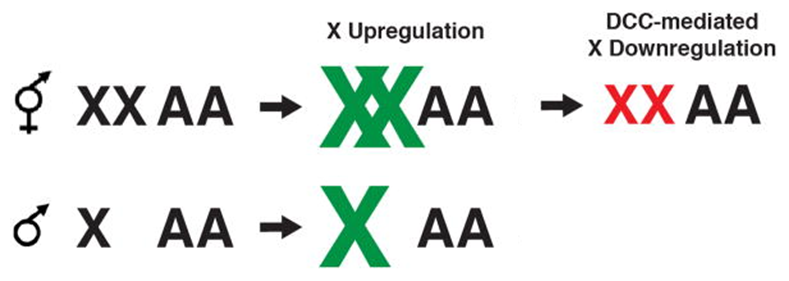
Upregulates males + hermaphrodites
Down regulate females
Ohno’s Hypothesis (1967)
X or Z linked genes will be expressed at twice the level of autosomes per active domain in the heterogametic sex
Wright and Mank PNAS 2012
How are mammalian sex chromosomes regulated?
Therian (non-egg laying) mammals
X chromosome inactivation in females
Xmale=Xfemale
Xmale= ? Automale
Xfemale= ? Autofemale
When did X-inactivation evolve?
Reviewed in Graves 2016
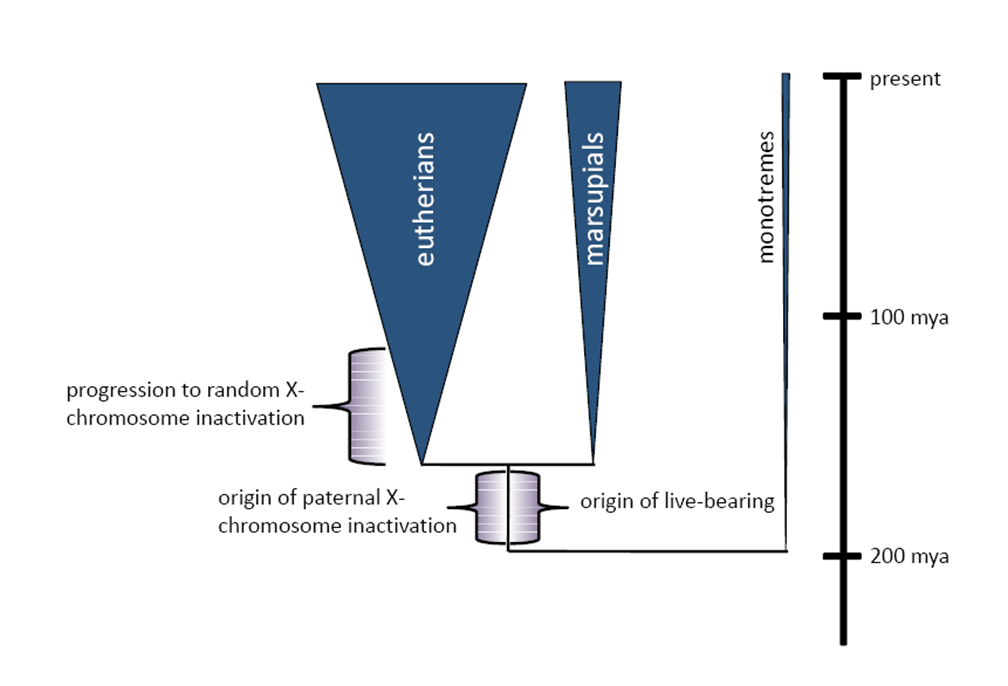
Incomplete/gene by gene dosage compensation
Studies across a broader range of taxa suggest that complete dosage compensation is the exception rather than the rule (monotreme mammals, birds, snakes, frogs, fish and plants).
Not all sex-linked genes are similarly sensitive to dose. Lowly expressed genes less sensitive, retained orthologues more sensitive
Complete dosage compensation more common in XY systems.
Telomeres
Repetitive nucleotide sequence at ends of chromosomes
A and T rich
Shorten at each cell division
Length inversely correlated with ageing
Protect genes near the ends of chromosomes from deterioration
What are germline cells protected from?
shortening by enzyme telomerase
Centromeres
No defined sequence
Role is to link sister chromatids (chromosome arms) in cell division
Attach to spindle fibres during meiosis
Role in enabling disjunction - chromosome segregation - when cell divides to generate normal chromosomal complement
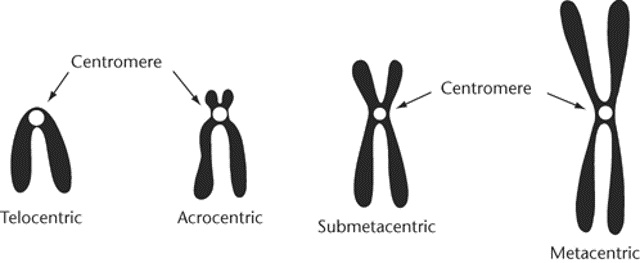
General rule of gene action + chromosome location
In multicellular organisms, evolution + gene action are independent of chromosome location
Exception to general rule of gene action + chromosome location: Position Effects in Hox Genes
HOX genes in plants and animals affect patterning of early embryos.
Mutations in HOX genes cause transformation of one body part into another.
HOX genes are clustered together.
Remarkably, their sequence along the chromosome corresponds to the order of segments in the body that they control.
Given the complexity of the interdependent developmental processes that they control, their co-location enables efficient regulation.
Exception to general rule of gene action + chromosome location: Tight Linkage
Selective forces acting on one gene can affect neighbours.
Consider nucleotide diversity within a region of sequence.
Plotted against distance to important site known to be under strong selective pressure.
At that site, diversity drops because one favoured variant is selected. Also, the
“well of diversity” effect extends to closely linked loci.
Chromosomes as units of evolution
Can act as genetic elements
Selection on a chromosome acts on 100s - 1000s of genes at a time
•The evolutionary effect on whole chromosomes depends on chromosome size – larger chromosomes typically (though not always) carry more genes.
Example of larger chromosomes carrying more genes
Drosophila melanogaster

Karyotype
Chromosomal genotypes
Karyotype also means the numbers of chromosomes in a haploid (1n) or diploid (2n) cell
Karyotype + chromosome number vary across species
Chromosome Mutations
Polyploidy – multiplication of the entire karyotype (e.g from 2n to 4n)
Autopolyploidy
Allopolyploidy
Chromosomal rearrangements
Translocation
Fusions
Fission
Inversions
Polyploidy
doubling of chromosome number
known as Paleopolyploidy in eukaryotes
Common in plants, less so in animals, some cases in fish + amphibians + also in fungi
What is polyploidy caused by?
nondisjunction during meiosis
a pair of homologous chromosomes has failed to separate or segregate at anaphase so that both chromosomes of the pair pass to the same daughter cell
autopolyploidy = both sets of chromosomes from a single specie s
Why are autopolyploids selected for in domesticated plants?
produce more protein per cell due to doubled dose of genes
Examples of domesticated autopolyploids
potato
banana
coffee
peanut
Why are polyploids reproductively isolated from parental diploid species?
due to the formation of triploids
unable to produce balanced gametes = sterile
speciation
Example: triploid sterile rainbow trout
Many fishery-raised fish populations are triploid (easy to generate: apply pressure to eggs, induces non-disjunction & bigger).
Allows for stocking for sport fishing, but prevents released fish from breeding with local populations and introducing non-native genetic diversity.
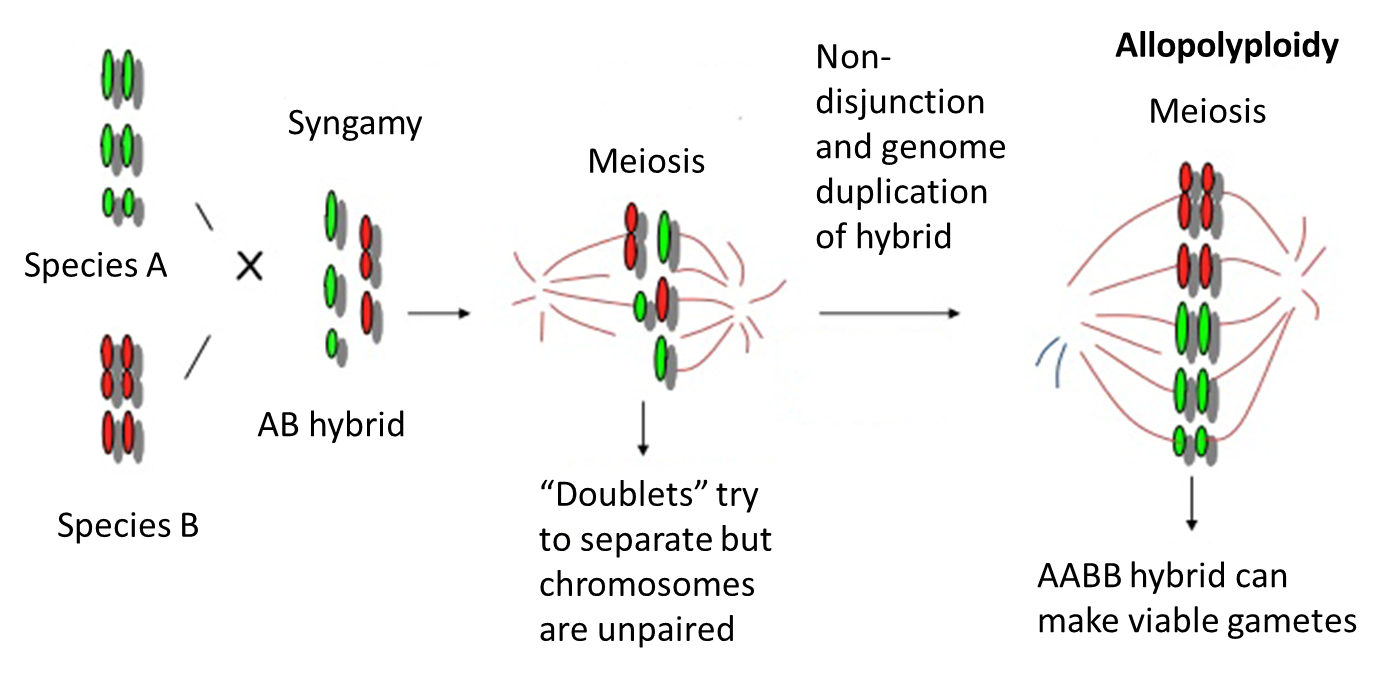
Allopolyploidy
two sets of chromosomes
each from a different parent species
result of hybridisation + nondisjunction
can make viable gametes
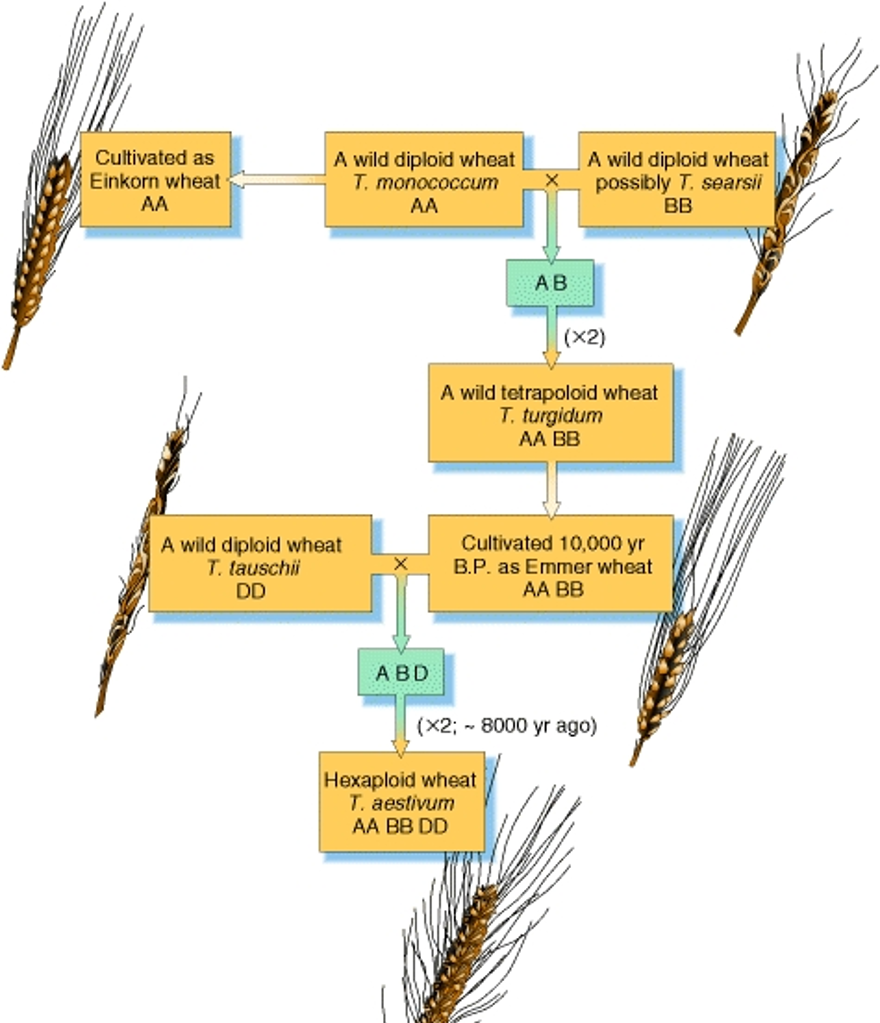
Allopolyploidy formation in Wheat, Triticum aestivum
Wheat = allohexaploid (2N chromosomes from each of 3 species) produced from two separate hybridisation events.
Each hybridisation was followed by chromosome doubling in the new hybrid; this enabled normal bivalent formation at meiosis and thus the production of fertile plants.
Genetic evidence suggests spontaneous hybridisation between T. urartu, einkorn wheat (A genome donor) and T. speltoides/searsii (B genome donor) created tetraploid species T. turgidum (emmer wheat).
Hexaploid wheat arose from second hybridisation between new tetraploid and another diploid, T. tauschii (D genome donor).
Chromosomal rearrangements
translocation
fusion
fission
inversion
segmental duplication
Reconstructing phylogenies using chromosomal movements
Take a species + isolate individual chromosomes on a basis of size/ weight
Search for areas of hybridisation to identify chromosomal movements during evolutionary time
Reconstruct phylogeny to common ancestor
Translocation
rearrangement between 2 non-homologous chromosomes
often result of incorrect chromosome pairing and recombination
no phenotypic consequence → still 2 copies of every gene
Example of translocation
“Philadelphia” karyotype
chronic myeloid leukaemia
translocation causes uncontrolled cell division
Chromosomal Fusions
When two chromosomes combine
More common in telocentric or acrocentric chromosomes due to centromere
Fusion of two telocentric chromosomes results in a metacentric chromosome with one functional centromere
Fusion of two metacentric chromosomes results in a dicentric chromosome (two functional centromeres). Can lead to chromosome breakage and sterility during meiosis.
Can be of evolutionary significance.
Example of chromosomal fusion
Human chromosome 2 is a result of the fusion of 2 acrocentric chromosomes after the last common ancestor with chimpanzees
Chromosomal Fissions
When a chromosome breaks in two
Technically possible, but far less common than fusion due to the need for a centromere
Without a centromere, a chromosome is not transmitted through meiosis, unable to attach to the spindle.
Other product has lost a chunk of material.
Usually deleterious and selected against.
Chromosomal inversions
Rearrangement where segment of chromosome rotated through 180 degrees
Homologous chromosomes can’t pair normally→ need to form a loop
Recombination between normal and inverted forms generate many gametes with incomplete complements, missing some vital genes
Consequences of crossing over within inversions
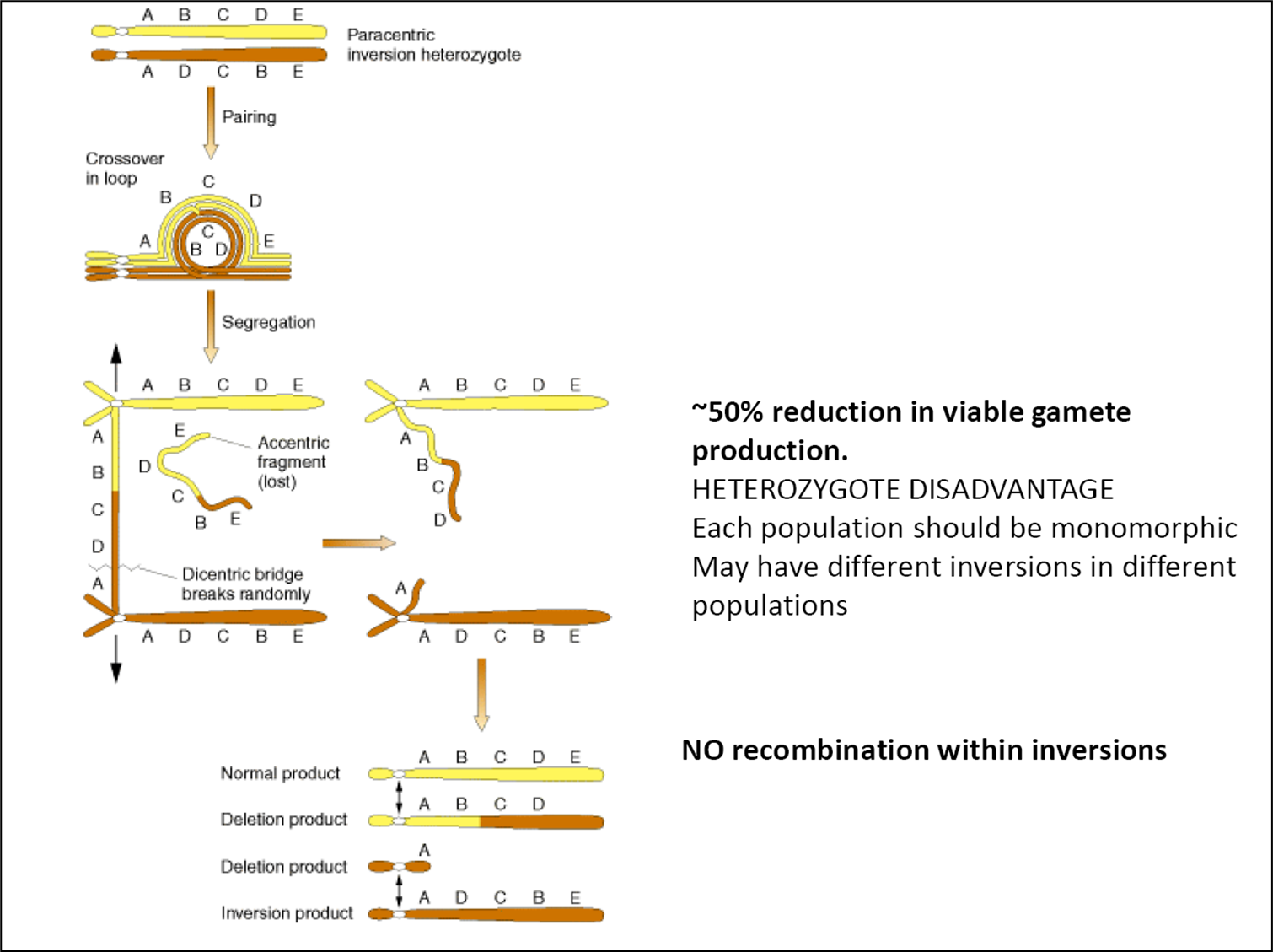
In this example:
2 out of 4 gamete types are fine
1 has lost 1 of the 5 gene regions
1 has lost most of its genes
Within a population, there will be selection against recombination within inversion
Adaptive inversions
Different inversions can arise in each population. Could allow local adaptation.
Due to suppressed recombination, an inversion might capture 2 or more alleles that are adapted to local environmental conditions and spread due to its selective advantage
Inversions might be adaptive sometimes because the suppression of recombination can link co-adapted alleles at nearby genes.
Genes that function in a related way can become strongly linked→ a “supergene”
Fire ant social forms: Monogyne
Workers tolerate only one queen → execute any others
New colony founded by new queen immediately following mating fight
extensive dispersal
Queen phenotype: larger, higher fecundity
2 copies of non-inverted SB/SB
Fire ant social forms: Polygyne
workers tolerate multiple queens
new colonies of queens and workers bud from existing colonies
dispersal not far
queen phenotype: smaller, lower fecundity
1 non-inverted + 1 inverted SB/Sb
No recombination.
Sb/Sb non-viable.
Adaptive inversions in Fire Ants -Wang et al. Nature 2013
Suppression of recombination between inversions can link co-adapted alleles at nearby genes
Monogyne & polygyne phenotypes are composed of multiple genes.
Phenotypes underpinned by 2 divergent forms of a chromosome (SB and Sb).
Region of chromosome with complete suppression of recombination between SB and Sb.
Study showed that different phenotypes map to a large inversion.
Evidence that genomic rearrangements maintain divergent phenotypes via local limits on recombination.
Segmental Duplications
Result from incorrect pairing and recombination
Half of resulting gametes have 2 copies of region (deleterious if any
genes are dosage sensitive)
Half of resulting gametes lack region (often lethal)
Bias towards smaller segmental duplications → less likely to have deleterious consequences
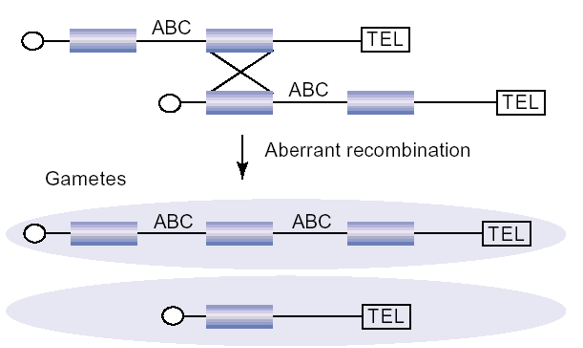
Example of Segmental Duplication: COWS
Normal beef breed = Aberdeen Angus
Belgian blue (double dose of myosin gene)
huge extra musculature
segmental duplication spans myosin gene
increased copy number of myosin gene I
increased production of myosin protein
into muscle formation