1404- Biochem- nitrogen metabolism- protein turnover
1/35
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
36 Terms
Essential amino acids importance
Cannot be synthesized by the body and must be obtained from the diet. They are crucial for protein synthesis, tissue repair, and metabolic functions.
Ubiquitin-proteasome system (UPS)
The UPS is the main pathway for targeted protein degradation in eukaryotic cells. It involves tagging proteins with ubiquitin and degrading them in the proteasome to regulate protein turnover and remove damaged proteins.
Role of ubiquitin in protein degradation
Ubiquitin serves as a molecular tag that marks proteins for degradation. At least four ubiquitin molecules must attach to a lysine residue on a target protein to signal its destruction in the proteasome.
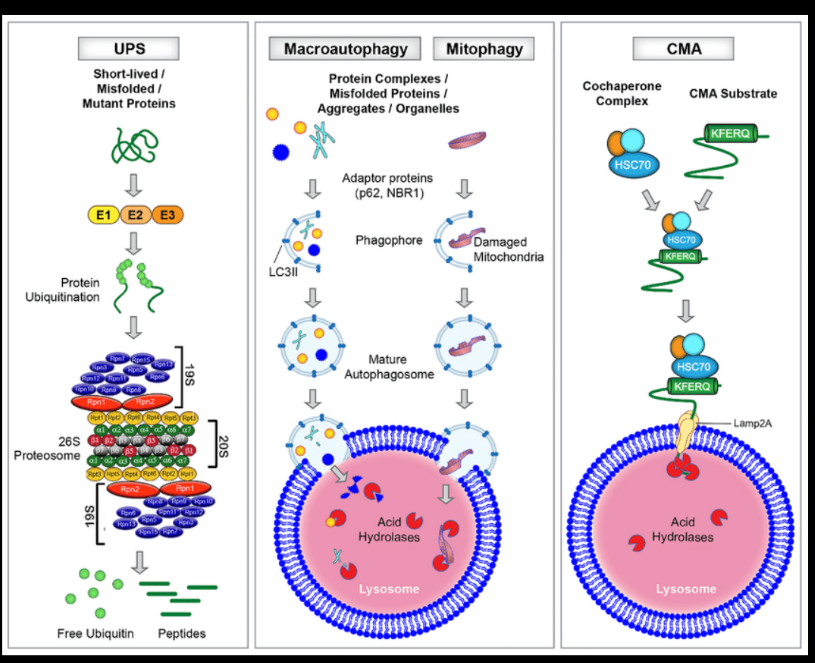
Autophagy vs. UPS
Autophagy degrades bulk cytoplasmic material and organelles through lysosomes, while the UPS selectively degrades short-lived proteins via proteasomes. Autophagy is more involved in cellular stress responses.
Chaperone-mediated autophagy (CMA) and key proteins
CMA selectively degrades proteins with a KFERQ-like motif. Key proteins: HSC70 (binds target proteins) and LAMP2A (lysosomal receptor for protein translocation).
Nitrogen balance and its significance
Nitrogen balance reflects the difference between nitrogen intake and nitrogen excretion. Positive balance (intake > output) occurs in growth and pregnancy, while negative balance (output > intake) indicates muscle wasting or malnutrition.
Estimating nitrogen balance
Estimated by dividing daily protein intake (g) by 6.25. Primary nitrogen output component: Urinary urea nitrogen (UUN), measured in 24-hour urine samples.

Three factors affecting nitrogen balance
1. Dietary intake (protein/energy availability), 2. Physiological states (growth, pregnancy, illness), 3. Disease/trauma (catabolic states increase nitrogen loss).
Marasmus vs. Kwashiorkor
Marasmus: Total calorie deficiency → muscle and fat wasting. Kwashiorkor: Protein deficiency with sufficient calories → edema and fatty liver.
Disease linked to UPS dysfunction
Angelman syndrome: Caused by mutations in the E6-AP ubiquitin ligase (UBE3A), leading to neurodevelopmental impairments.
Endogenous proteins
Proteins made within the body
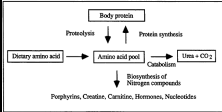
Importance of dietary protein
Needed to synthesize some endogenous proteins
Body cannot synthesize certain amino acids called essential amino acids
Essential amino acids
Valine
tryptopan
threorine
leucine
lysine
Histadine
isoleucine
methione
phenylalanine
protein turn over
The balance between protein synthesis and degradation by the continual renewal or replacement of proteins.
Importance of protein turnover
To maintain optimal functioning of proteins
Factors influencing protein turn- over
Type of tissue
relative contribution of varying tissues
age
adaptation of various levels of protein in take
Steady state protein turn over
Protein synthesis and degradation rates are balanced
Constant protein levels within cells
Transcriptome and metablome are not affected
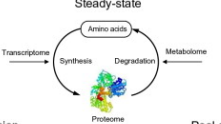
Transcriptome
Total amount of mRNA molecules expressed from a gene of an organism.
Metablome
Total number of metabolites within a system
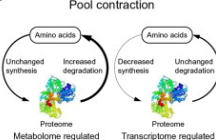
Amino acid pool expansion
Increased synthesis of proteome, unchanged degradation of proteome to amino acid→ transcriptome regulated
Unchanged synthesis of proteome, decreased degradation of proteome to amino acid→ metablome regulated
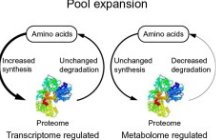
Amino acid pool contraction
Unchanged synthesis + decreased degradation of proteome→ Metablome regulation
Decreased synthesis + unchanged degradation → Transcriptome regulation
Protein degradation pathways
Ubiquitin- proteasome system
Macro autophagy/ Mitophagy
Chaperone mediated autophagy
Ubiquitin- proteasome system (UPS)- second definition
A major selective pathway for the degradation of misfolded, damaged or unneeded proteins to maintain homeostasis within eukaryotic cells
Step 1 of Ubiquitin - proteasome system
E1- Ubiquiting activating enzyme activate Ubiquiting in an ATP dependent manner.
Proteostasis network
The collective processes that maintain the stability and functionality of the proteome, ensuring that proteins are correctly folded, assembled, and degraded as necessary.
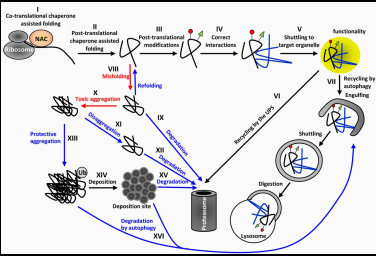
Step 1 of proteostasis network: Protein Synthesis
Proteins are synthesized from amino acids based on the instructions coded in the DNA.
Step 2 of proteostasis network: Protein Folding
Newly synthesized proteins must fold into their specific three-dimensional structures, facilitated by molecular chaperones.
Step 3 of proteostasis network: Quality Control
Misfolded or unfolded proteins are identified and either refolded or targeted for degradation.
Step 4 of proteostasis network: Protein Assembly
Proteins may need to assemble into larger complexes, requiring precise interactions with other proteins.
Step 5 of proteostasis network: Protein Turnover
Proteins have a lifespan and are continuously degraded and replaced; this balance is crucial for cellular health.
Step 6 of proteostasis network: Response to Stress
Cells activate protective mechanisms in response to stress conditions to maintain proteostasis.
Step 2 of Ubiquitin - proteasome system
E2- Ubiquiting conjugating enzyme transfers activated Ubiquiting with the help of E3
Step 3 of Ubiquitin - proteasome system
E3 facilitates the transfer of Ubiquitin from E2 to the lysine residue on the target cell
Step 4 of Ubiquitin - proteasome system
The first Ubiquitin attaches to the amino acid of the side chain of a lysine residue on the isopeptide bond
Step 5 of Ubiquitin - proteasome system
Additional ubiquitins are added to form a multi-ubiquitin chain which acts as a marker for proteasome to recognize.
Steps 6 of Ubiquitin - proteasome system
Proteosome recognized marker, targets protein, degrades it into peptides and also degrades the ubiquitins to be reused