Molecular Genetics
1/79
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
80 Terms
What are the components of nucleic acids?
Nucleic acids are macromolecules composed of three main components: a five-carbon sugar, a phosphate group, and a nitrogenous base
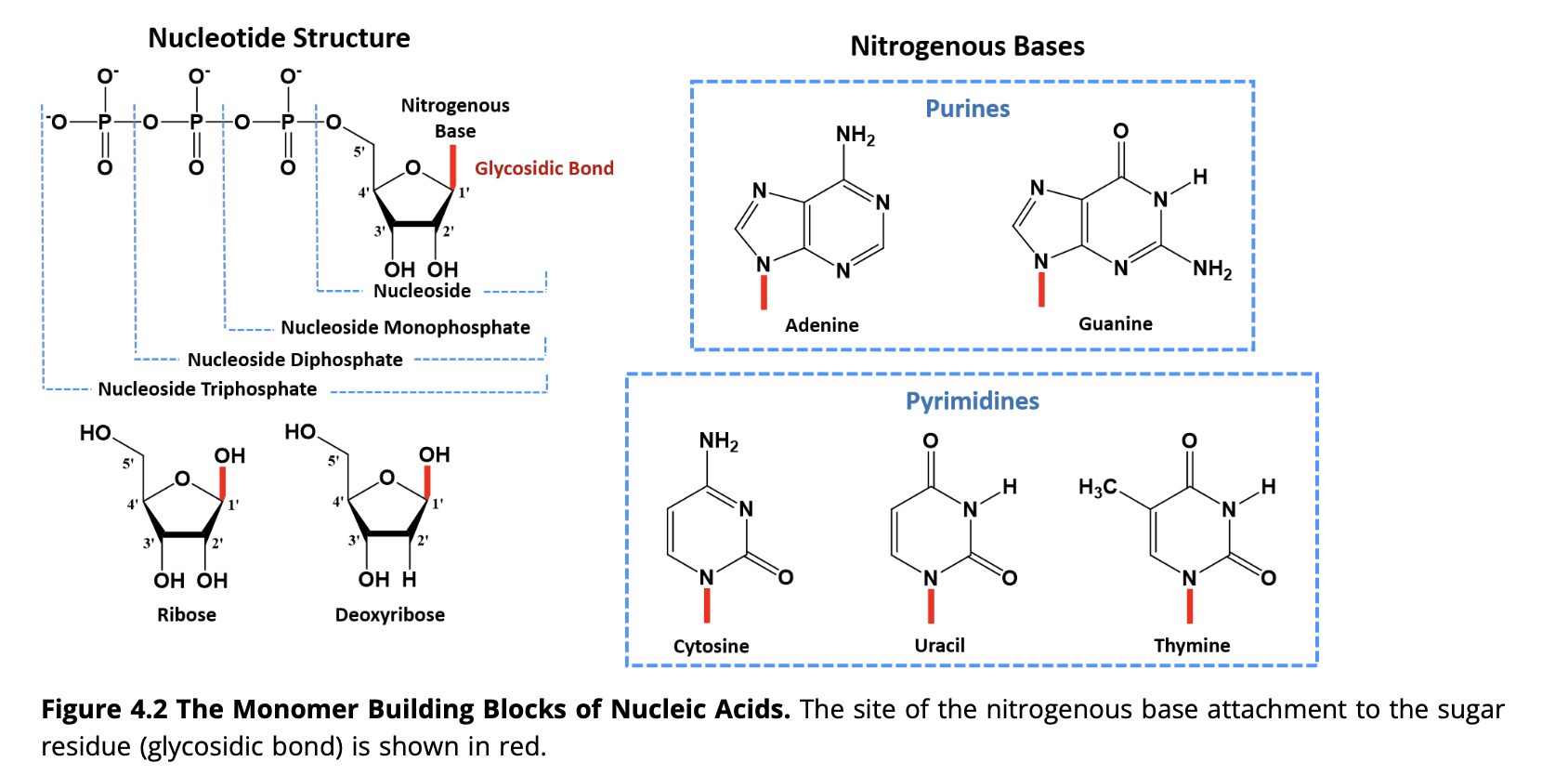
What types of bonds are present in DNA?
DNA contains hydrogen bonds between base pairs and phosphodiester bonds between sugar and phosphate groups.
Tell me more about phosphodiester bonds
They are covalent bonds between the phosphate group on the 5’ carbon of one nucleotide to the hydroxyl group on the 3’ carbon of the next nucleotide. This creates the alternating sugar-phosphate backbone of DNA and RNA.
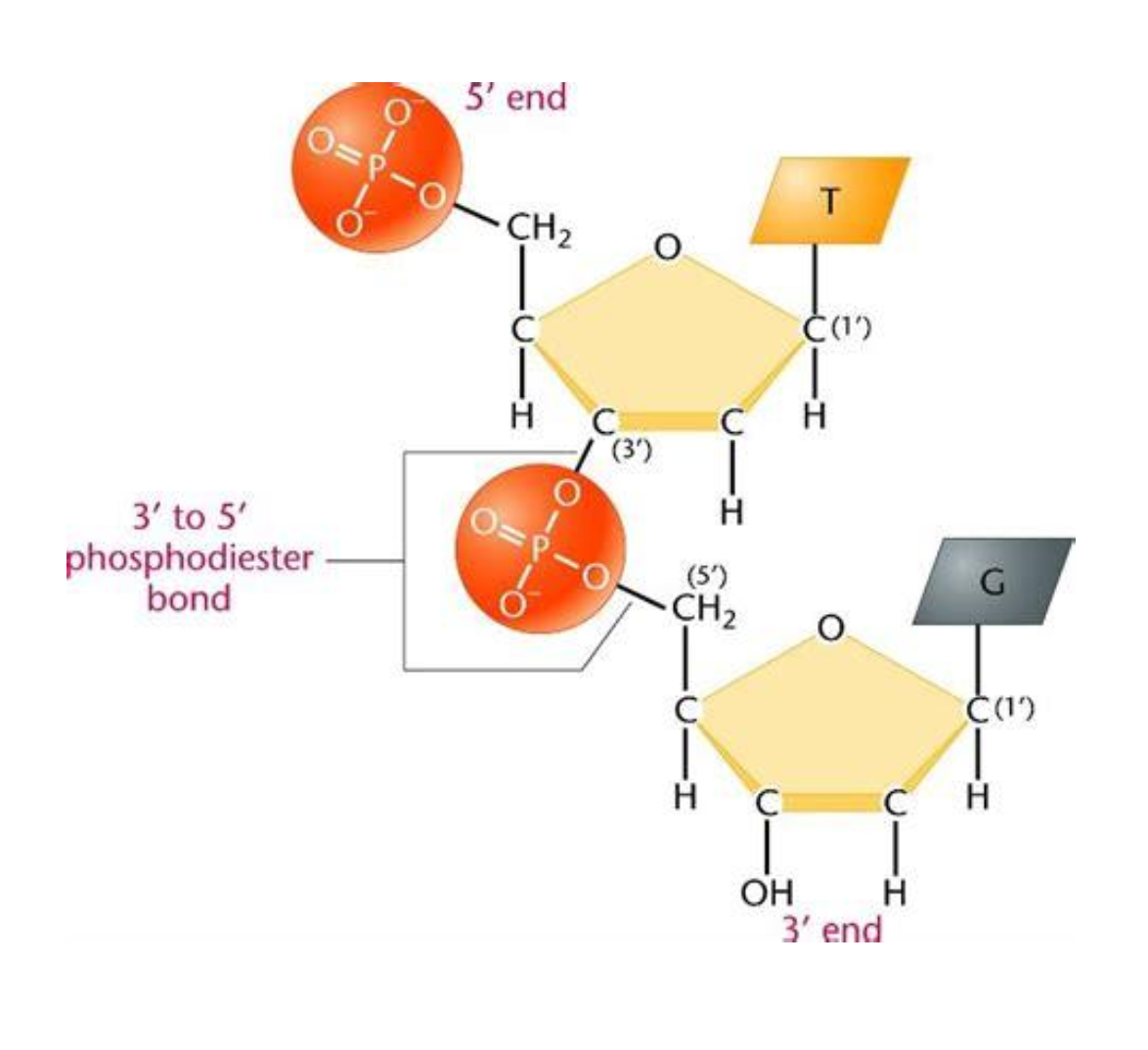
What bases bond together to give complimentary base pairing?
Adenine pairs with Thymine, and Cytosine pairs with Guanine. In RNA, Adenine pairs with Uracil instead of Thymine.
How many hydrogen bonds between base pairs?
Adenine and Thymine (or uracil) form two hydrogen bonds, while Cytosine and Guanine form three hydrogen bonds.
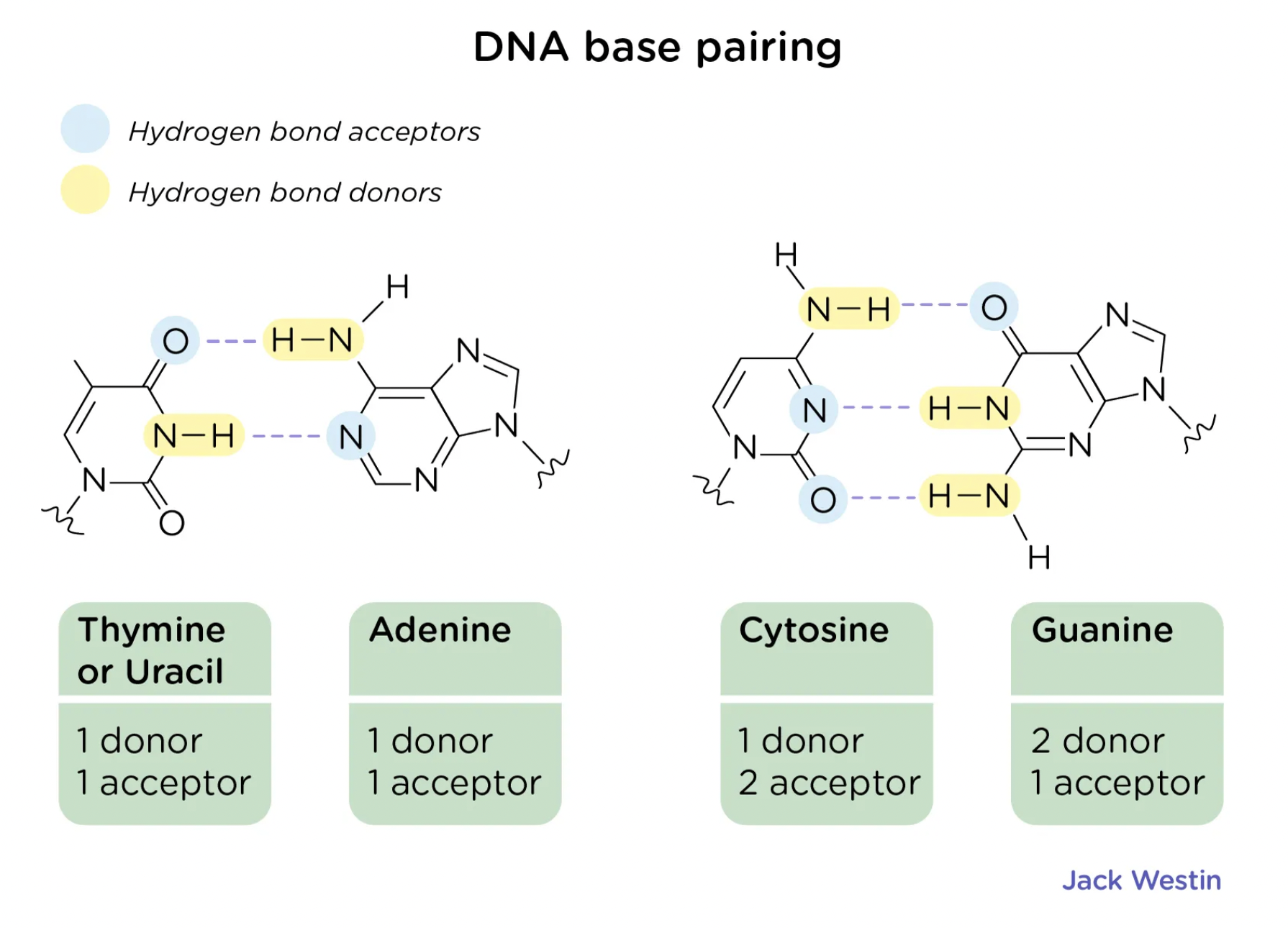
How does hydrogen bonding affect the structure of DNA
Hydrogen bonds are individually weak enough to allow for easy separation of strands during transcription and replication but are collectively strong enough to maintain structural integrity and stability of DNA.
They provide stability and specificity in base pairing - contributing to the helical shape of DNA.
What direction do the strands of DNA run?
They run in an antiparallel arrangement. One strand runs 5’ to 3’ and the other runs 3’ to 5’. This antiparallel arrangement is essential for complimentary base pairing and DNA replication. It provides structural stability and symmetry to the helix.
How does the major and minor grooves in DNA helix shape form?
Bases attach asymmetrically to the backbone - exposing different edges of base pairs. The uneven attachment causes the backbone to be closer together on one side, forming the minor groove and further apart on the other, creating the wider major groove.
What are the differences between the major and minor grooves?
The Major Groove is the primary site for sequence specific protein binding as it offers a more expansive and chemically diverse surface for sequence-specific recognition. Proteins such as helix-turn-helix (HTH) proteins and zinc finger proteins (ZNFs) may bind here.
Each base pair acts as a recognition code of functional groups. Proteins can distinguish all four base pairs because of the way the hydrogen bond - donor/acceptor - hydrogen atom and methyl group are attached to each other.
The Minor Groove has much less chemical diversity. Mainly hydrogen acceptors, donors and hydrogen atoms. Fewer molecules to attach to mainly due to the fact of much less specific discrimination of A & T and G & C. Therefore the minor groove becomes a target for small molecules and protein that sense the shape of DNA and the flexibility.
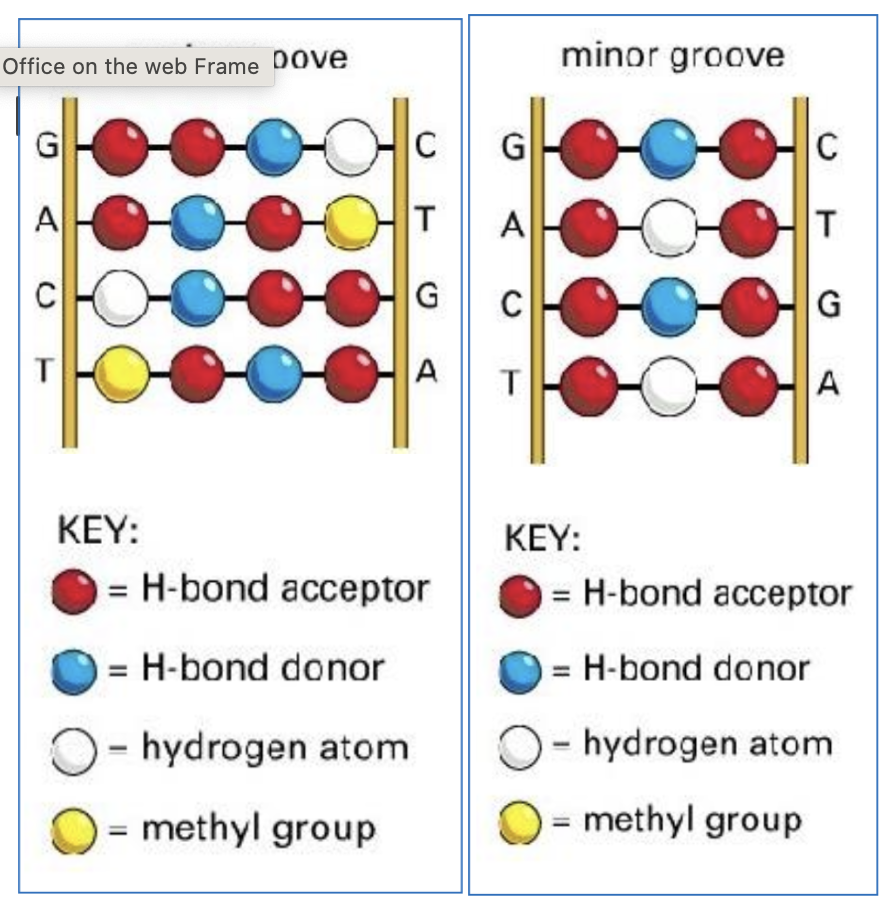
What size are the major and minor grooves?
major groove - 2.2nm
minor groove - 1.2nm
DNA helix can be dynamic in shape. Name 3 alternative conformations.
B-DNA
Z-DNA
G4 Quadruplex
How does B-DNA conformation affect structure and stability?
Right handed helix
Most stable under physiological conditions
How does Z-DNA conformation and structure and stability?
Zig-Zag sugar phosphate backbone
GC rich sequences and negative supercoiling
Important in gene regulation and immune recognition
How does G4 conformation affect structure and stability?
Formed in G rich sequences
4 guanines form hoogsteen hydrogen bonded tetrads
important in gene stability, replication and transcription regulation
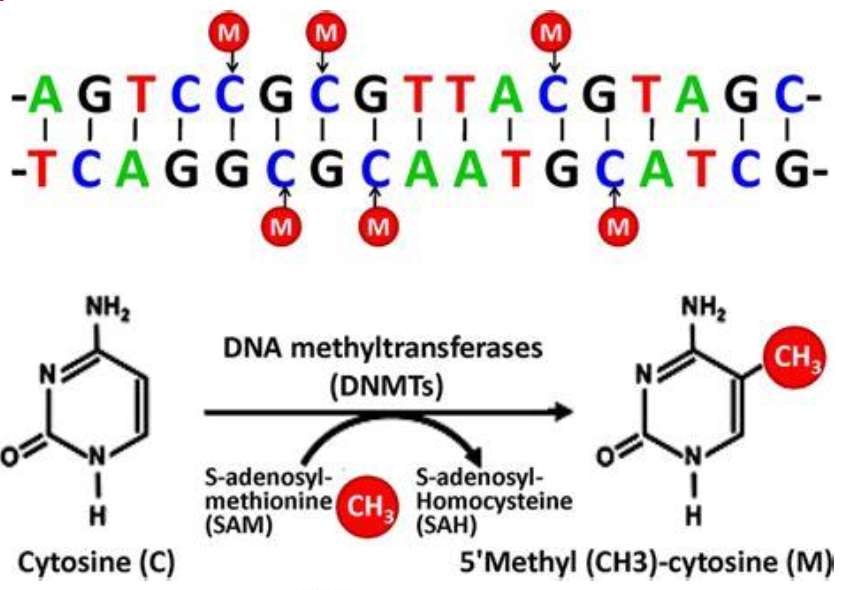
DNA methylation is a chemical modification of DNA. Tell me more about it.
5-Methylcytosine
Applied at the C5 position of cytosine by methyltransferases
Involved in epigenetic regulation
Stable but reversible mark, linking DNA sequence to expression
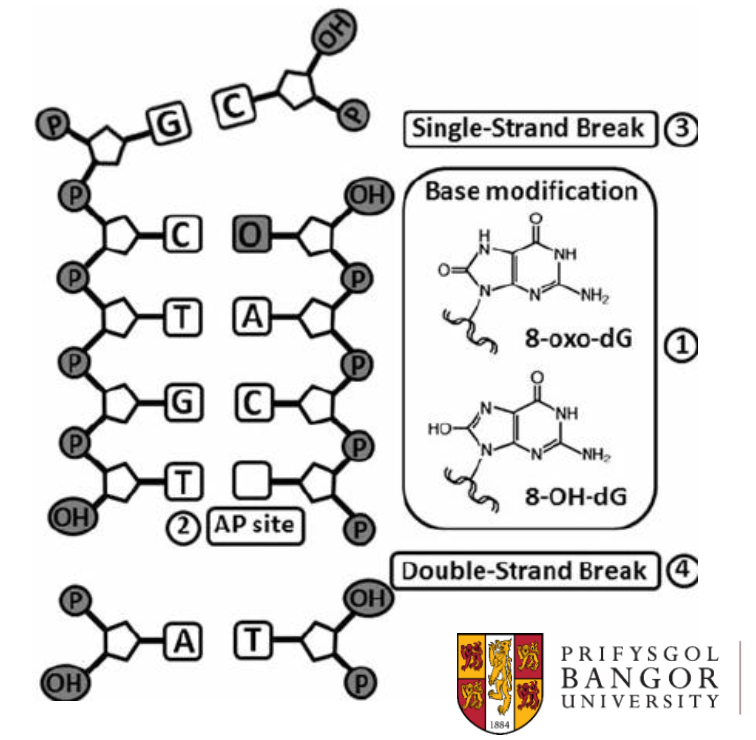
Oxidative base modifications are chemical modifications of DNA. Tell me more about them.
Caused by reactive oxygen species
8-oxoG mispairs with A
Causes G-T transversions
part of DNA damage process
Seen particularly in cellular senescence. When our cells get older, especially in cells with lots of mitochondria where lots of oxygenation is happening, more ROS will be present.
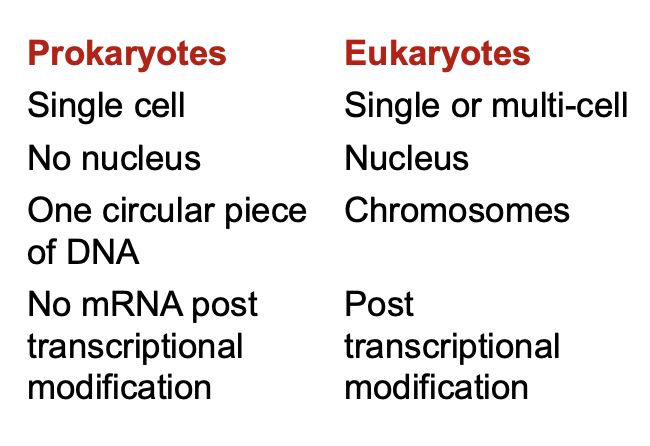
What are the differences in the Prokaryotic and Eukaryotic Genome?
How many base pairs in the human genome?
>3 billion
16.5k in mitochondrial DNA the rest (around 3.2bill) in nuclear DNA
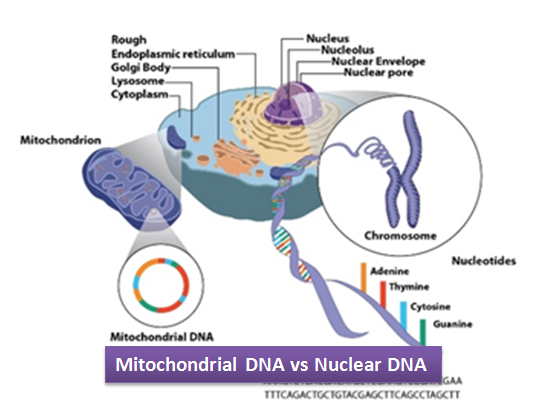
Where is the human genome stored?
In two organelles -
The Nucleus - one copy per cell
The mitochondria - multiple copies per cell
How many protein coding genes in the human genome?
20,000 - 30,000 (mostly nuclear - only 37 in mitochondrial DNA)
What percentage of inter-individual identity in the human genome?
99.6%
what percentage of the human genome is responsible for the differences between us?
<1%
When was the first complete sequence of the human genome published?
2022
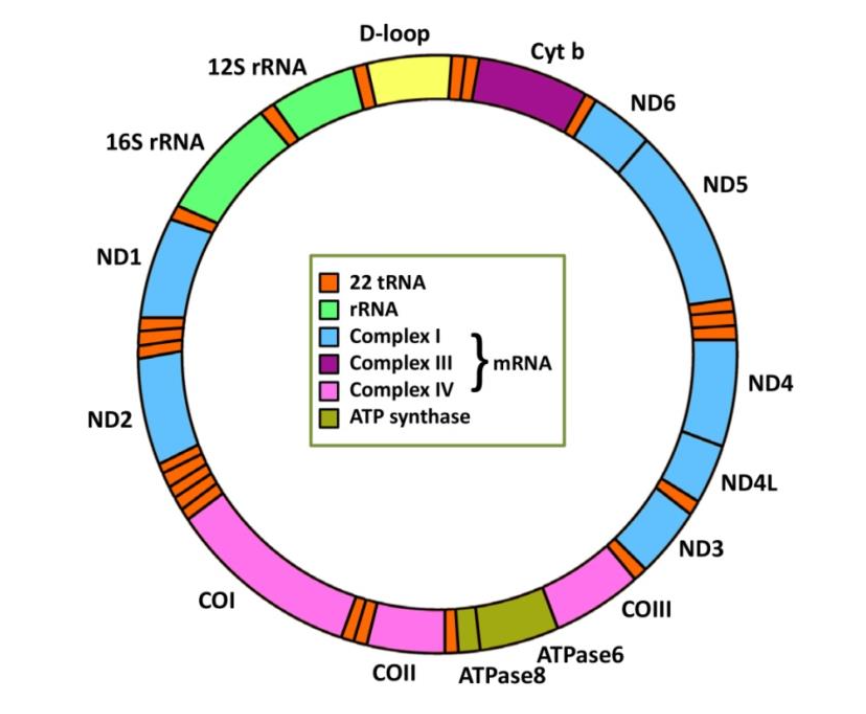
What shape is mitochondrial DNA (mtDNA)?
Double stranded circular DNA
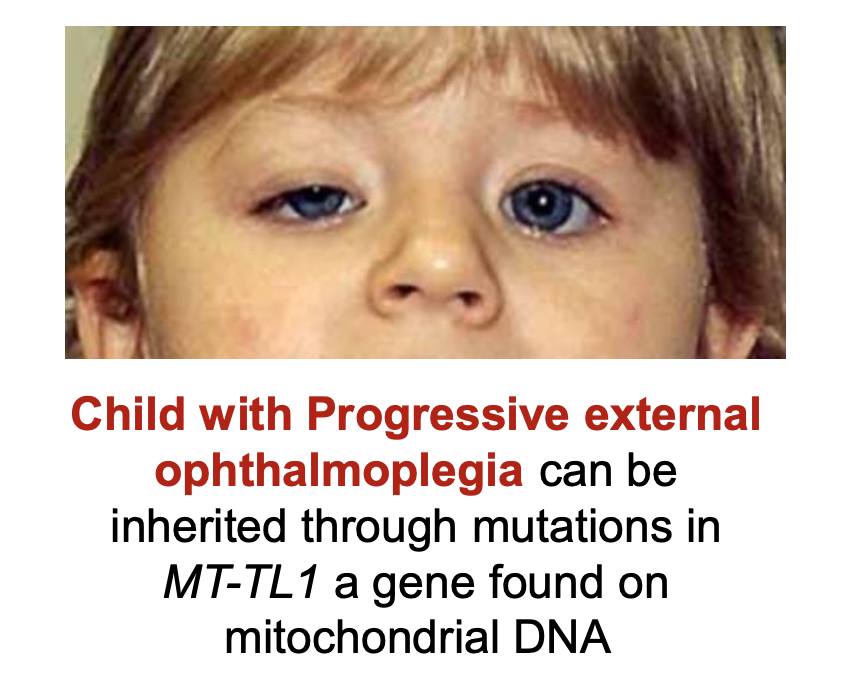
what are the differences in inheritance in nuclear and mtDNA?
nuclear = mendelian
mtDNA = maternal
Where does a higher rate of mutations in DNA occur
mtDNA
what does mtDNA code for?
mitochondrial ribosomes and transfer RNAs
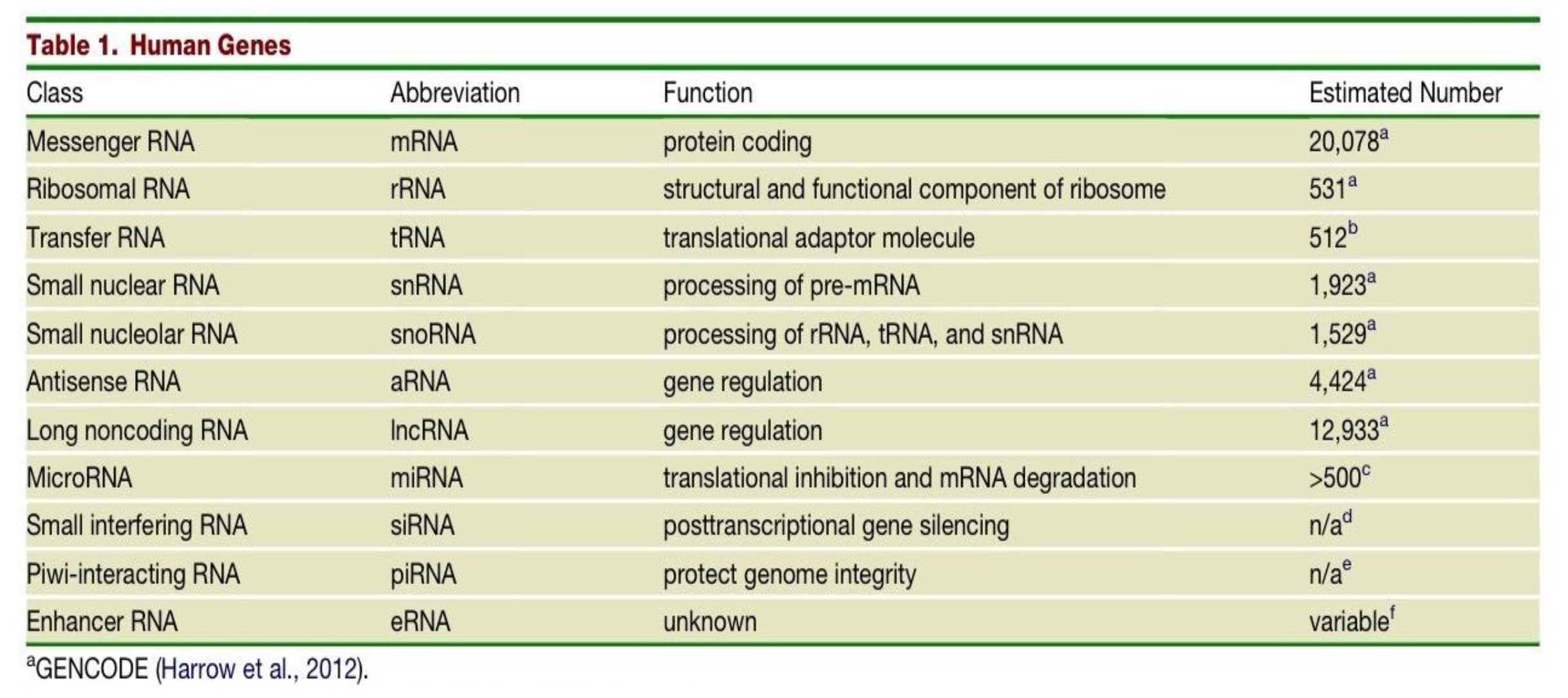
Which class of human genes have a protein coding function and what is their estimated number in the human genome?
messenger RNAs (mRNAs)
20,078
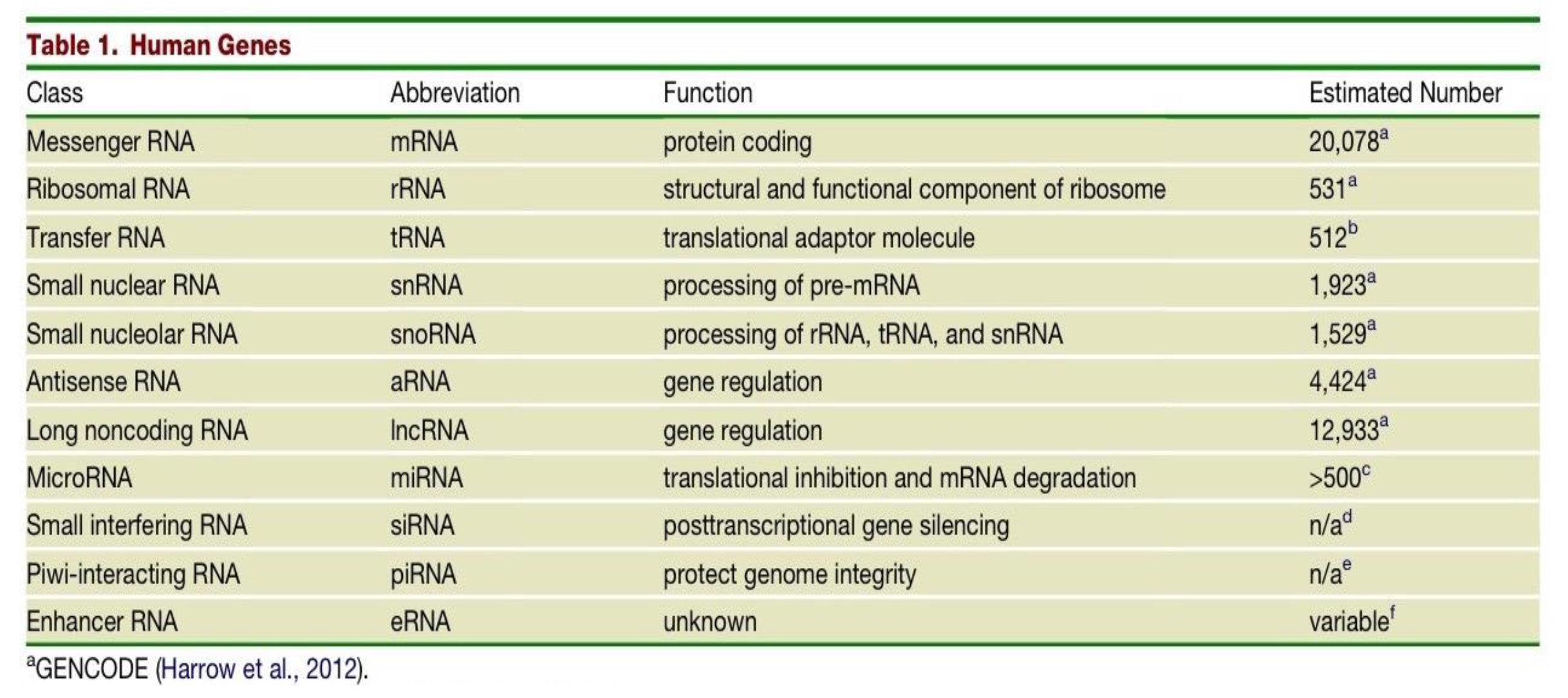
What is the function of the class of gene - Ribosomal RNA (rRNAs) and what is their estimated number in the human genome?
structural and functional component of the ribosome
531
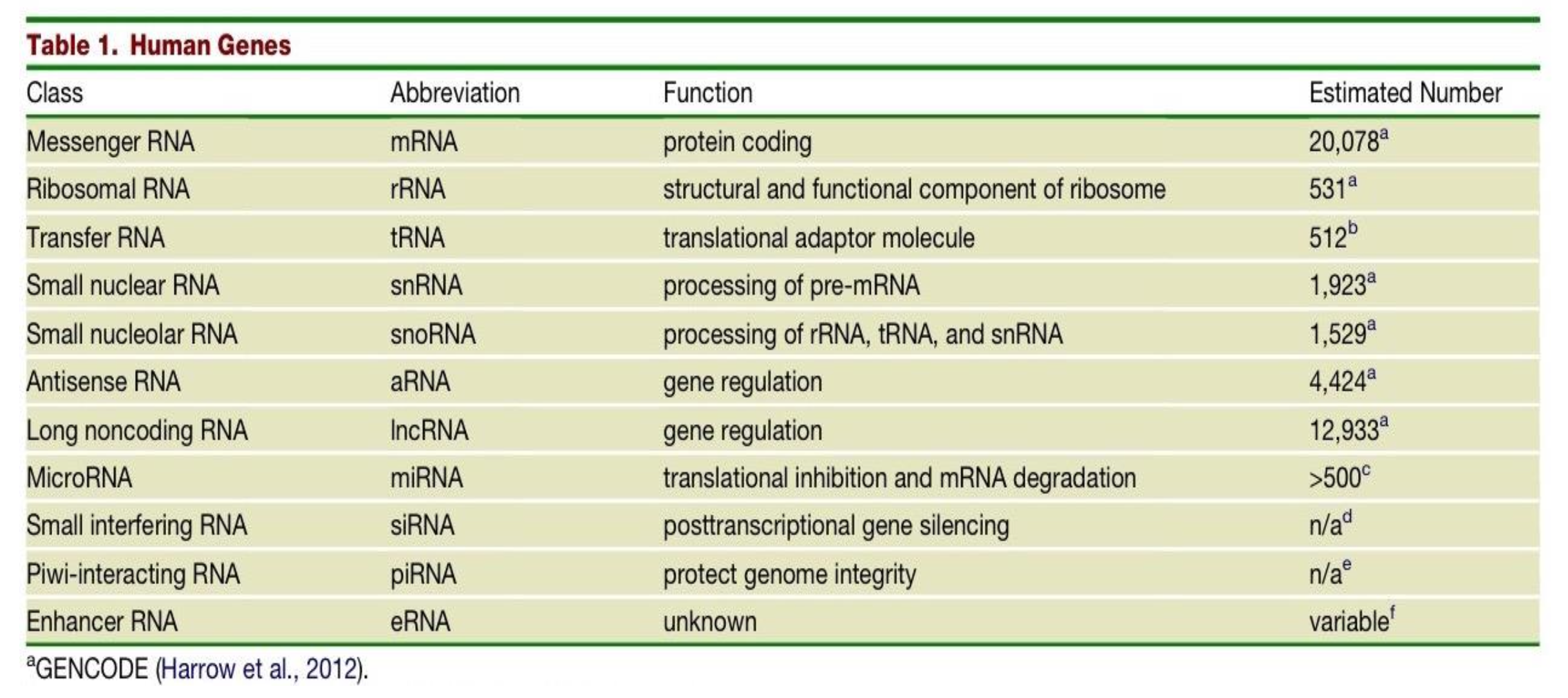
The estimated number of transfer RNAs (tRNAs) in the human genome is 512, what is their function?
they are translational adaptor molecules
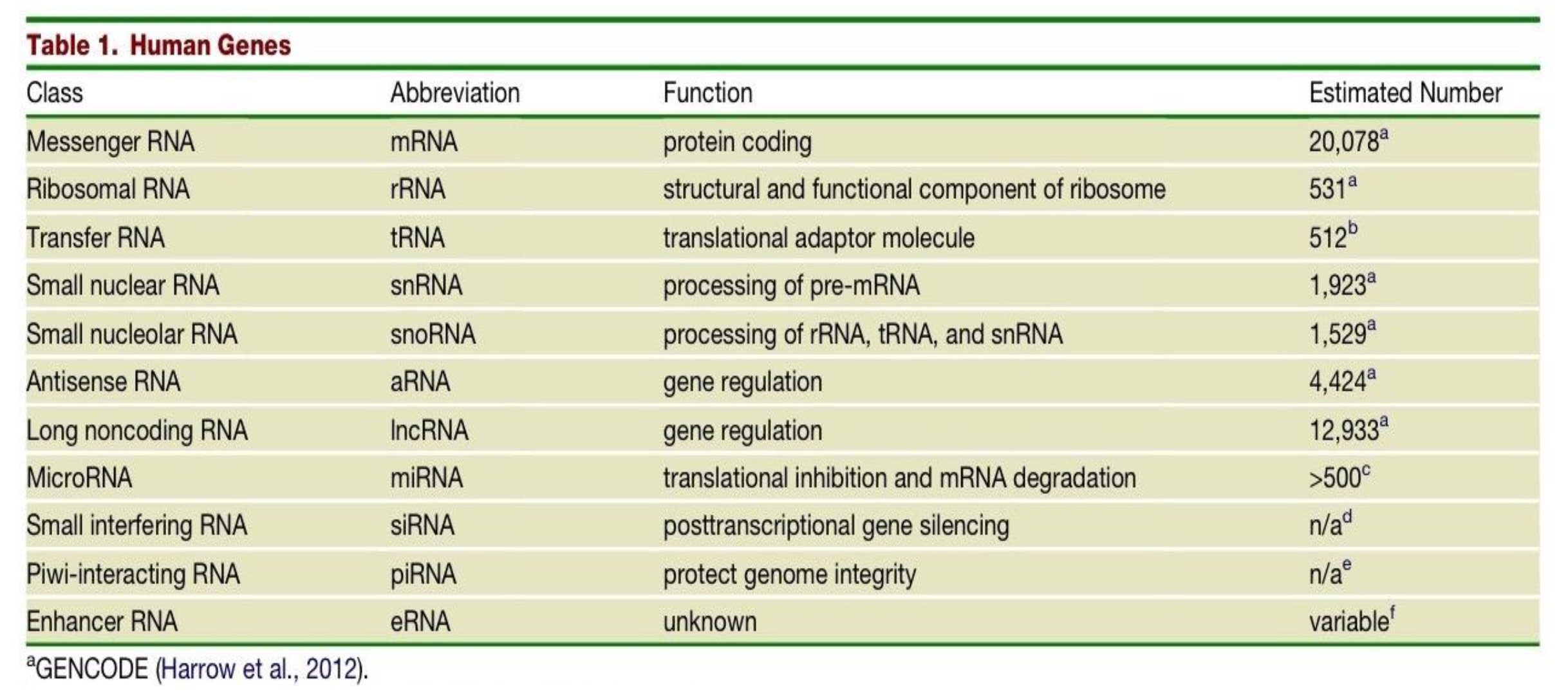
What are the differences in Small nuclear RNAs (snRNAs) and Small nucleolar RNAs (snoRNAs)?
snRNAs - function is processing of pre-mRNA. Estimated number in human genome is 1,923.
snoRNAs - function is processing of rRNA, tRNA and snRNA. Estimated number in human genome is 1,529.
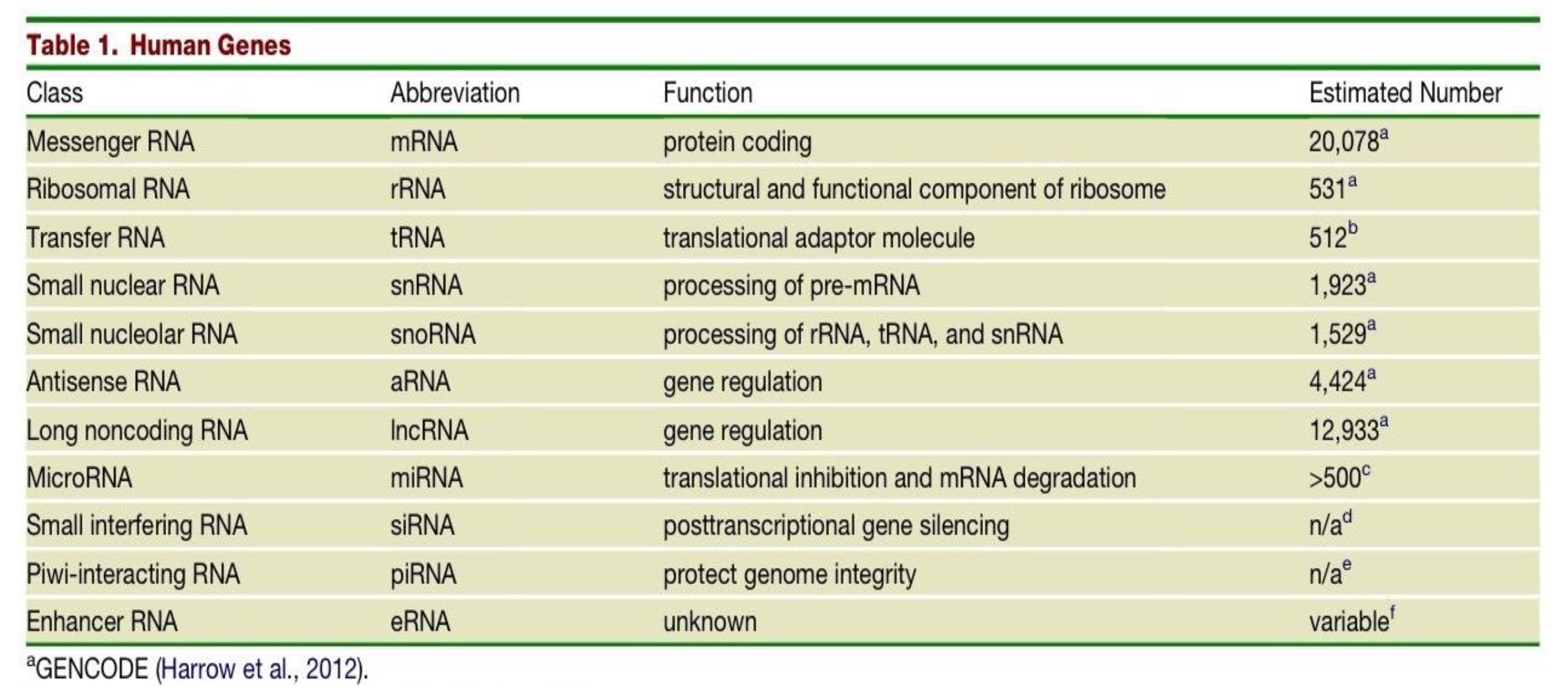
Two classes of genes in the human genome have the function of gene regulation - what are they and what are their respective estimated number in the human genome?
Antisense RNA (aRNA) - 4,424
Long noncoding RNA (lncRNA) - 12,933
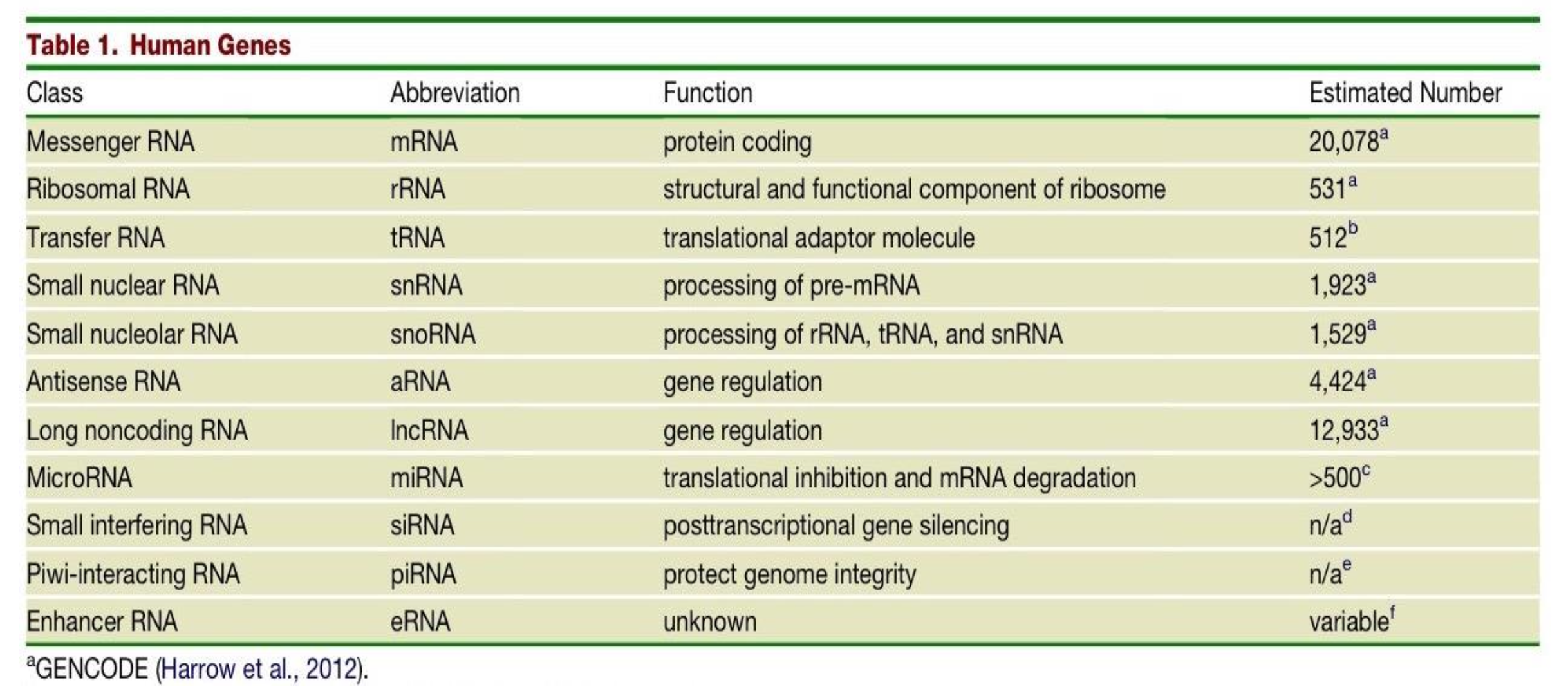
What is the function of MicroRNAs (miRNA) and what is their estimated number in the human genome?
translational inhibition & mRNA degradation
>500
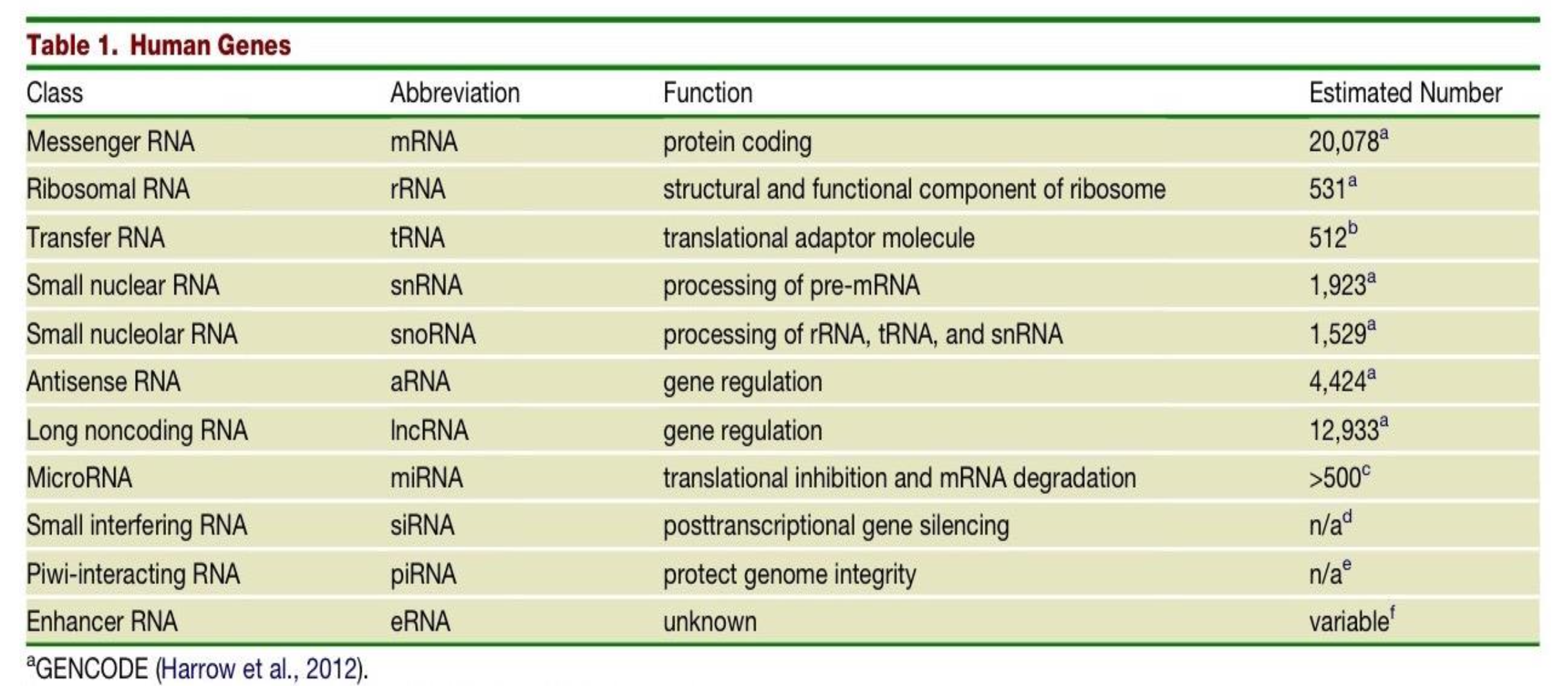
What is the function and estimated number in the genome of Small interfering RNAs (siRNAs)?
post transcriptional gene silencing
n/a
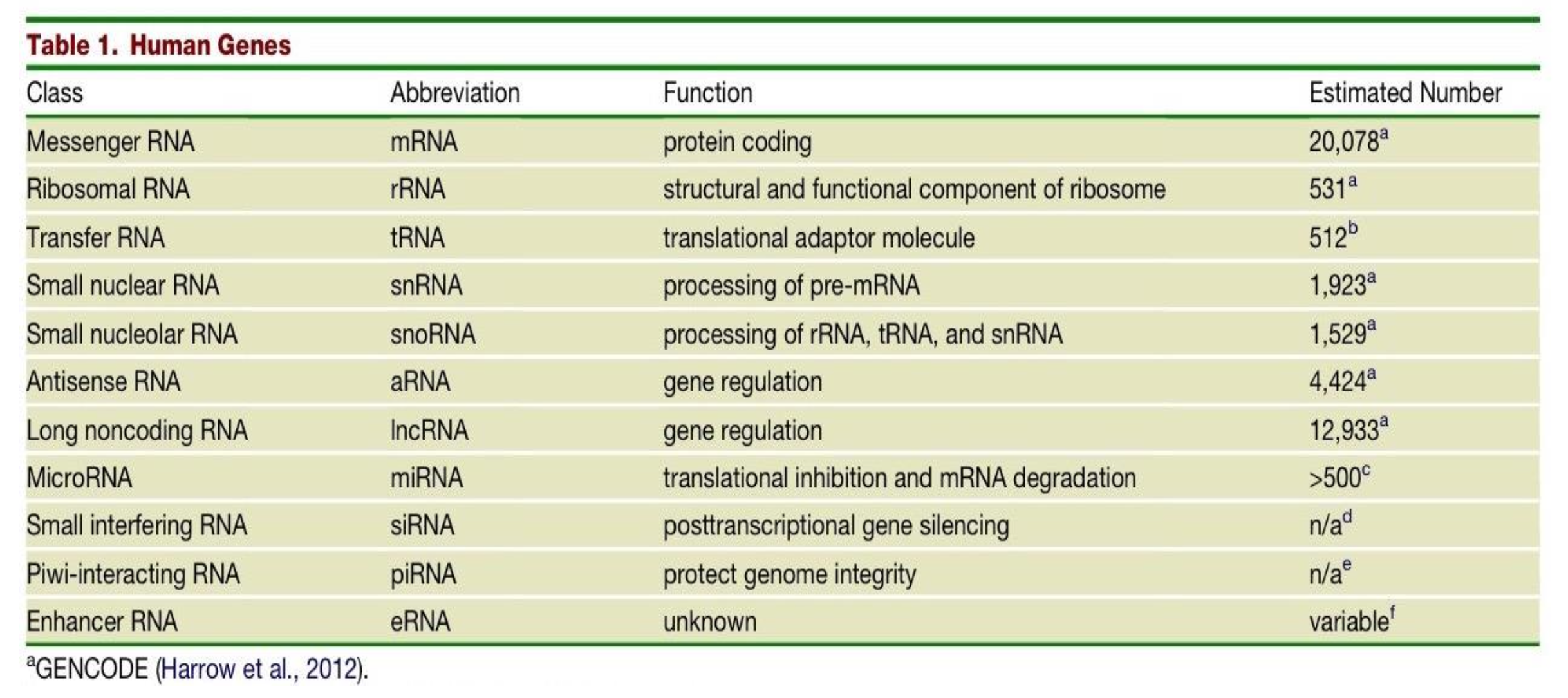
What is the function and estimated number in the genome of Piwi-interacting RNA (piRNA)?
protecting genome integrity
n/a
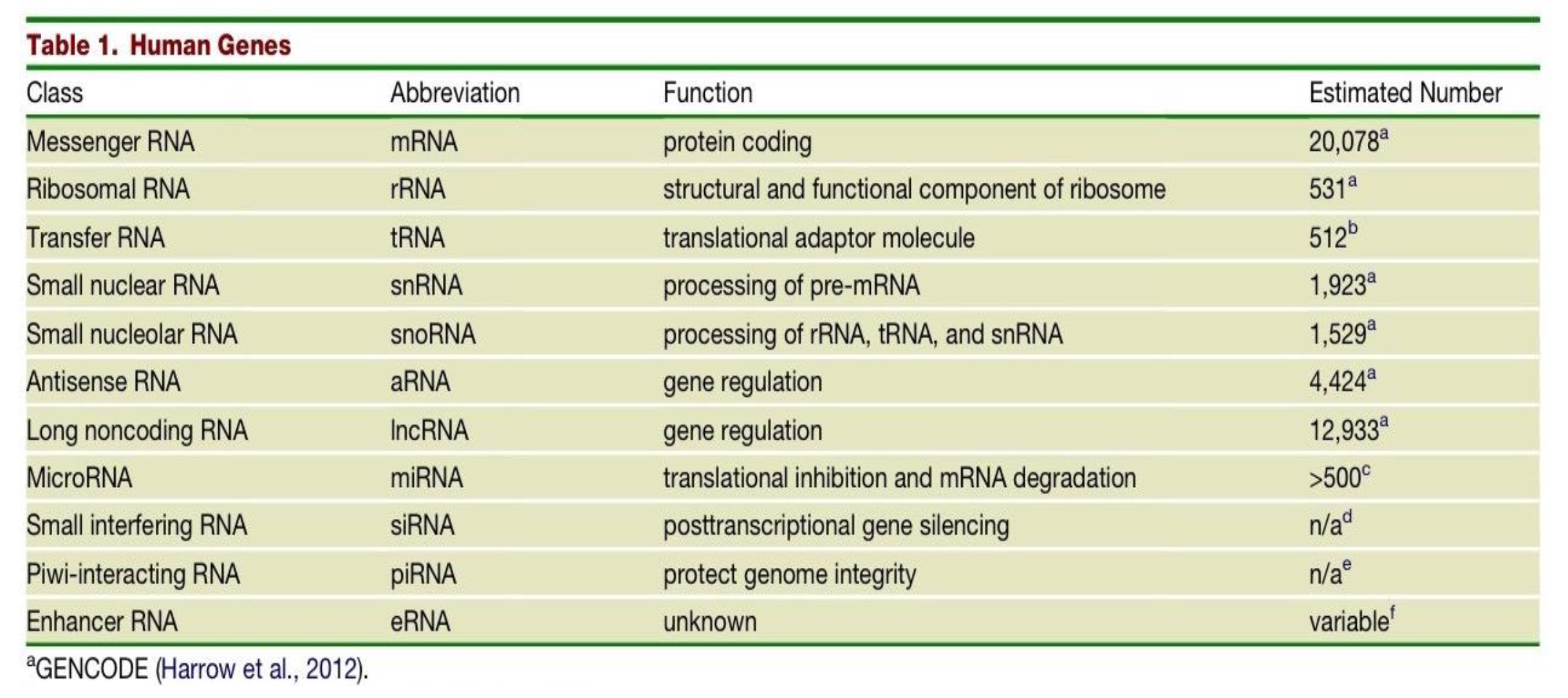
What is the function and estimated number in the genome of Enhancer RNA (eRNA)?
unknown function
variable number
How do protein coding genes become RNA in Eukaryotes?
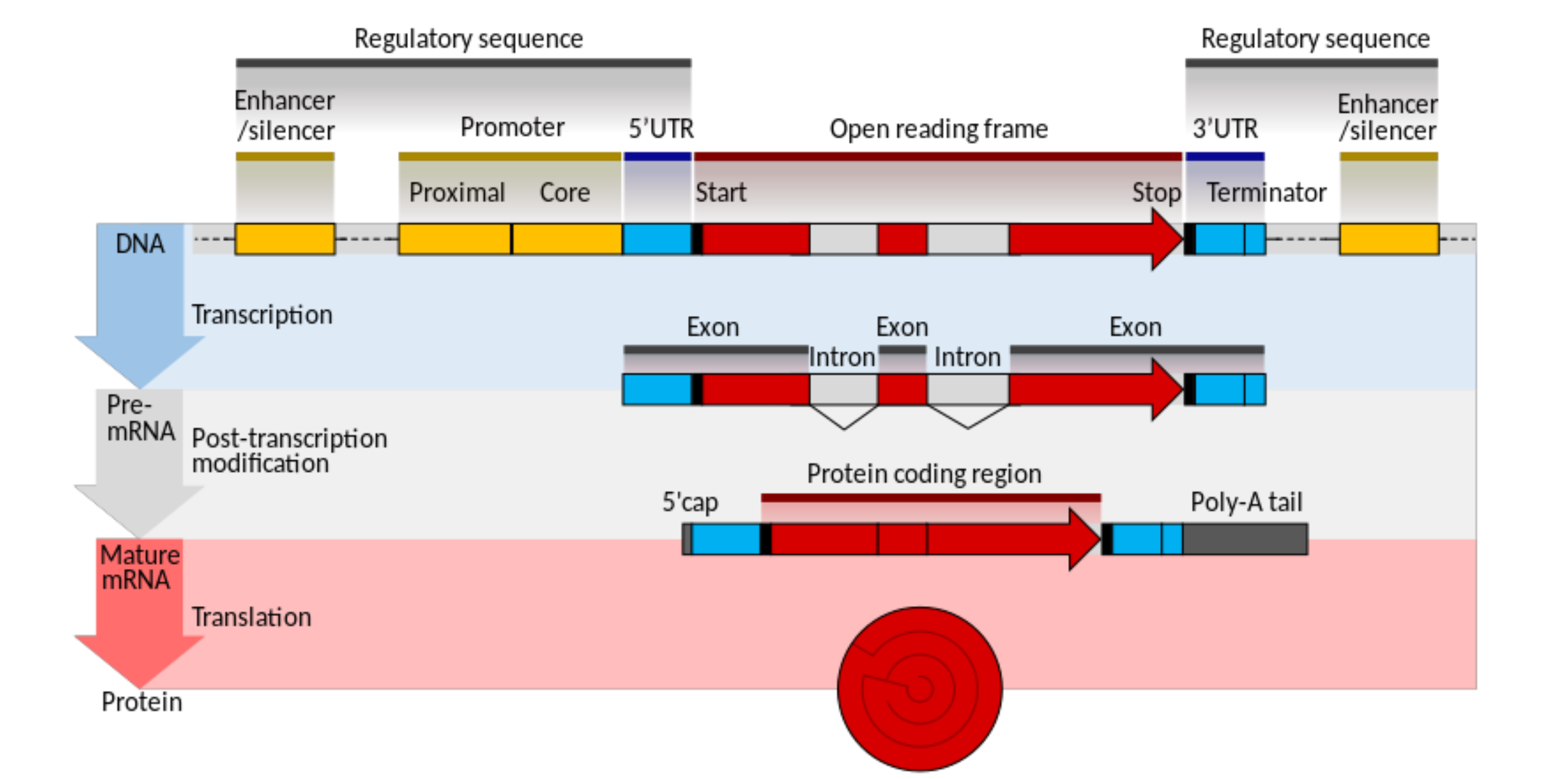
How do protein coding genes become RNA in Prokaryotes?
no pre-mRNA / post transcriptional modification of mRNA stage
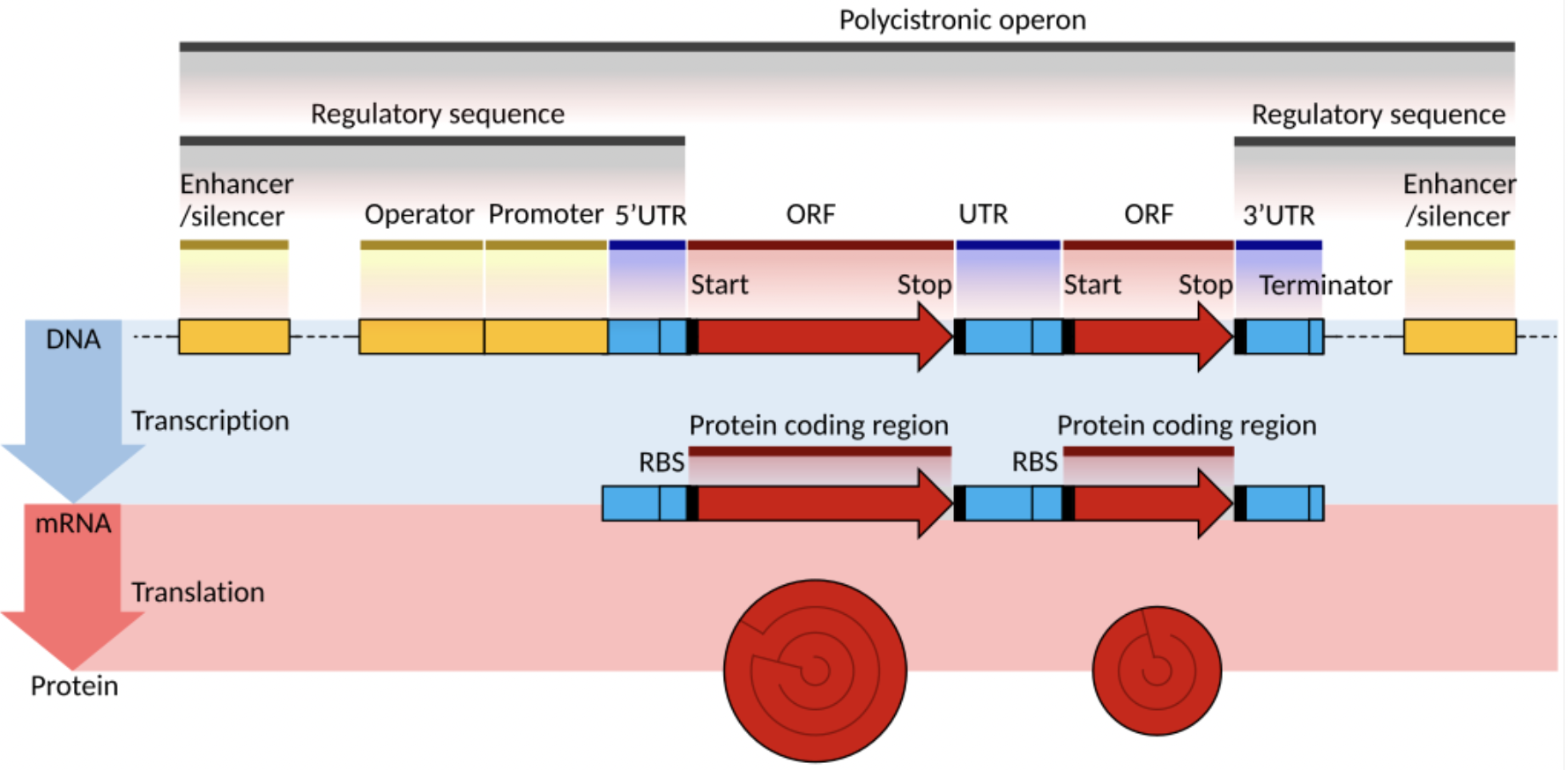
How can only 25,000 to 30,000 protein coding genes produce the massive variety of cells and tissues that exist in the human body?
alternative splicing
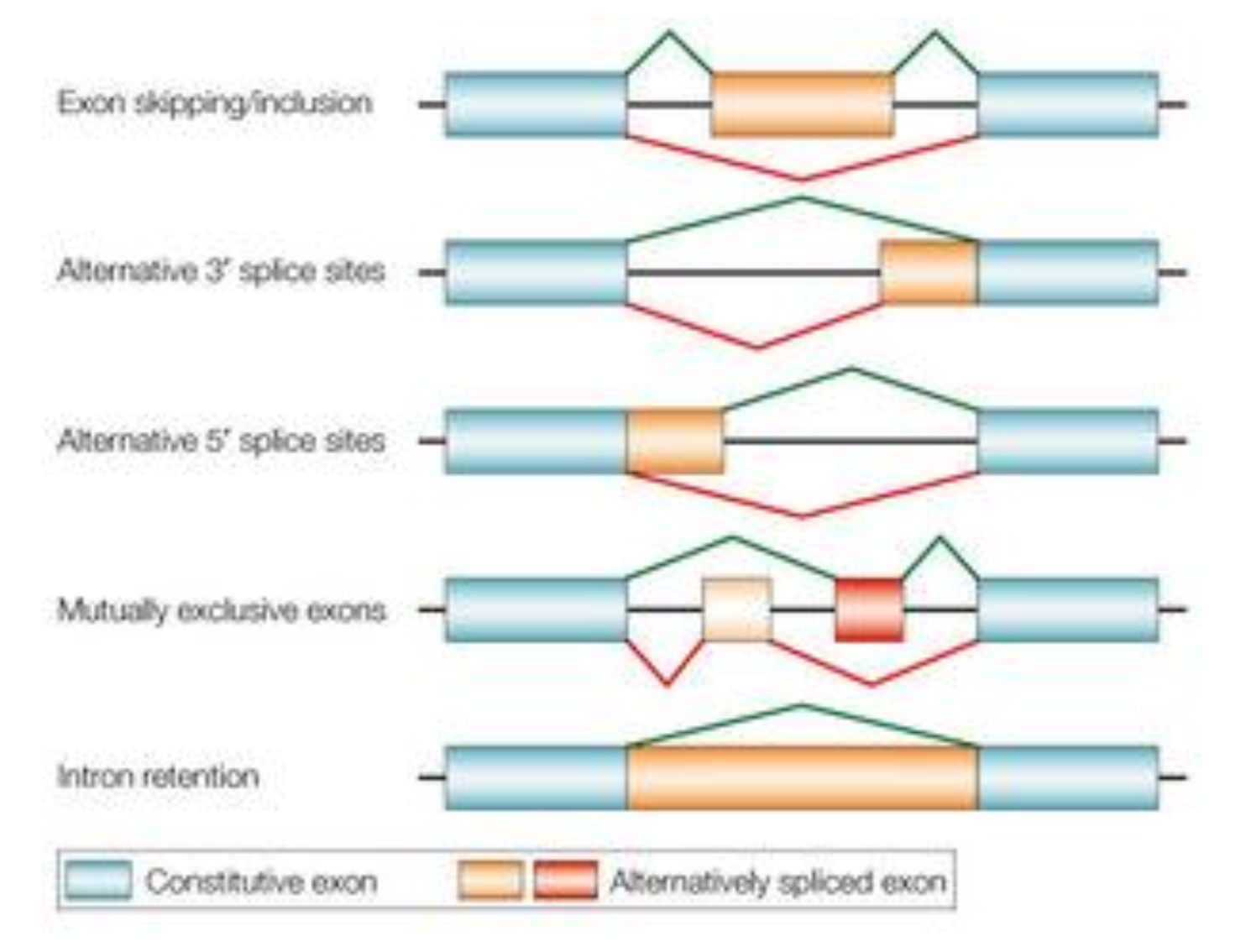
What are protein isoforms and how are they created?
Different proteins that come from the same gene.
Created by cutting out specific exons in a gene.
e.g. a gene has 15 exons (areas coding for protein) and we only need the first 5 to create a protein isoform. We splice the gene and remove the 10 exons not needed.
hundreds of protein isoforms can be created from a single gene by alternatively splicing the gene and selecting and discarding different variations of exons along the gene.
what percentage of the human genome is made up of non-coding DNA?
around 98%
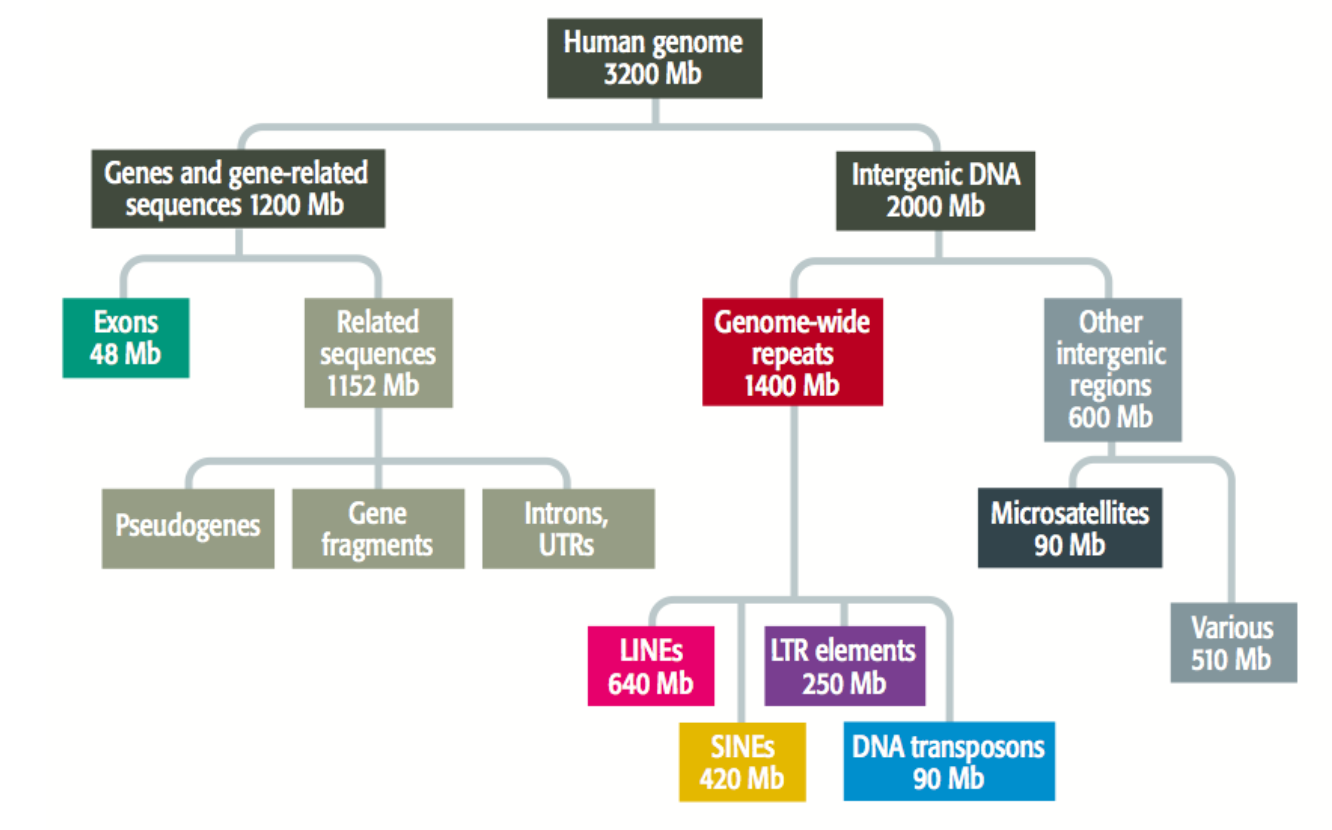
List three types of non-coding DNA:
Regulatory sequences
Non-coding RNAs
Repetitive elements
Non-coding DNA has a wide range of functions. List some:
Gene expression regulation
Maintaining chromosomal structure
Drive evolution
What are repetitive elements?
DNA with the same sequence e.g. ACTACTACTACT that are found in multiple copies across the genome.
What are some types of repetitive elements:
Tandem repeats: repeated DNA sequences found next to each other - broad spectrum of which satellites are a part of
Satellites: Large, highly repetitive sequences in centromeres (centre of DNA) or telomeres (end loops of DNA - help protect from digestive elements).
Mini and Microsatellites: Smaller repeats, often in telomeres or near genes.
Interspersed repeats: Transposable elements

Why are repetitive elements important?
For maintaining chromosomal structure and centromere function.
What are the most common motifs in microsatellites?
CA repeats or single nucleotide repeats e.g. AAAAA
Why are microsatellites often used in DNA fingerprinting?
No two individuals will have the same microsatellite profile. We all have different digestive enzymes which break the DNA apart into its barcode sequences - we can see variation is the length and the number of repeats an individual has and so we can use it as a way of identifying people. Microsatellites on the short arm of chromosome 6 are amplified and labeled using PCR.
What is Microsatellite instability (MSI)
The inability for your cells to keep your microsatellites at the same length when going through transcription. This can be a good indicator at how your cancer will progress and how well you will do when undergoing cancer treatment.
What are Retrotransposons?
Mobile elements propagated by RNA intermediates.
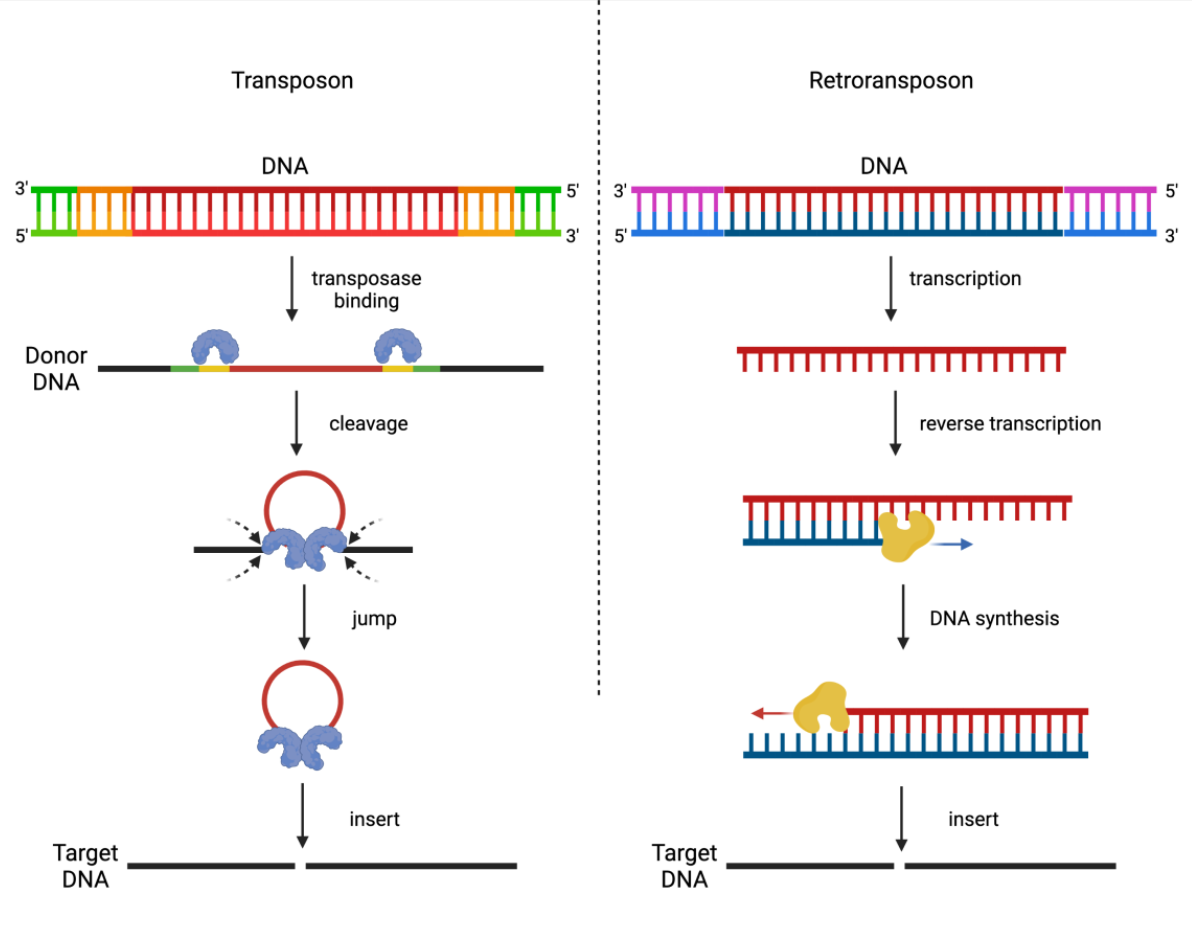
What do retrotransposons include?
LINES: Long Interspersed Nuclear Elements
SINES: Short Interspersed Nuclear Elements
LTR Retrotransposons: Mostly inactive
Tell me about LINES:
6-8kb encoding for proteins in retrotrasnposition (ORF1/ORF2)
LINE-1 (L1) accounts for about 17% but only has around 100 active copies
Involved in: Gene disruption, exon shuffling, structural variation
Tell me about SINEs:
100-300bp
Rely on LINE proteins
Alu elements: around 10% of the human genome
Involved in: alternative splicing and transcription interference
What is the functional role of transposons or TE-encoded transcripts in embryonic & somatic cells?
In Mouse LINE1 acts as a nuclear RNA scaffold for recruitment of key proteins required for maintaining stem cell identity
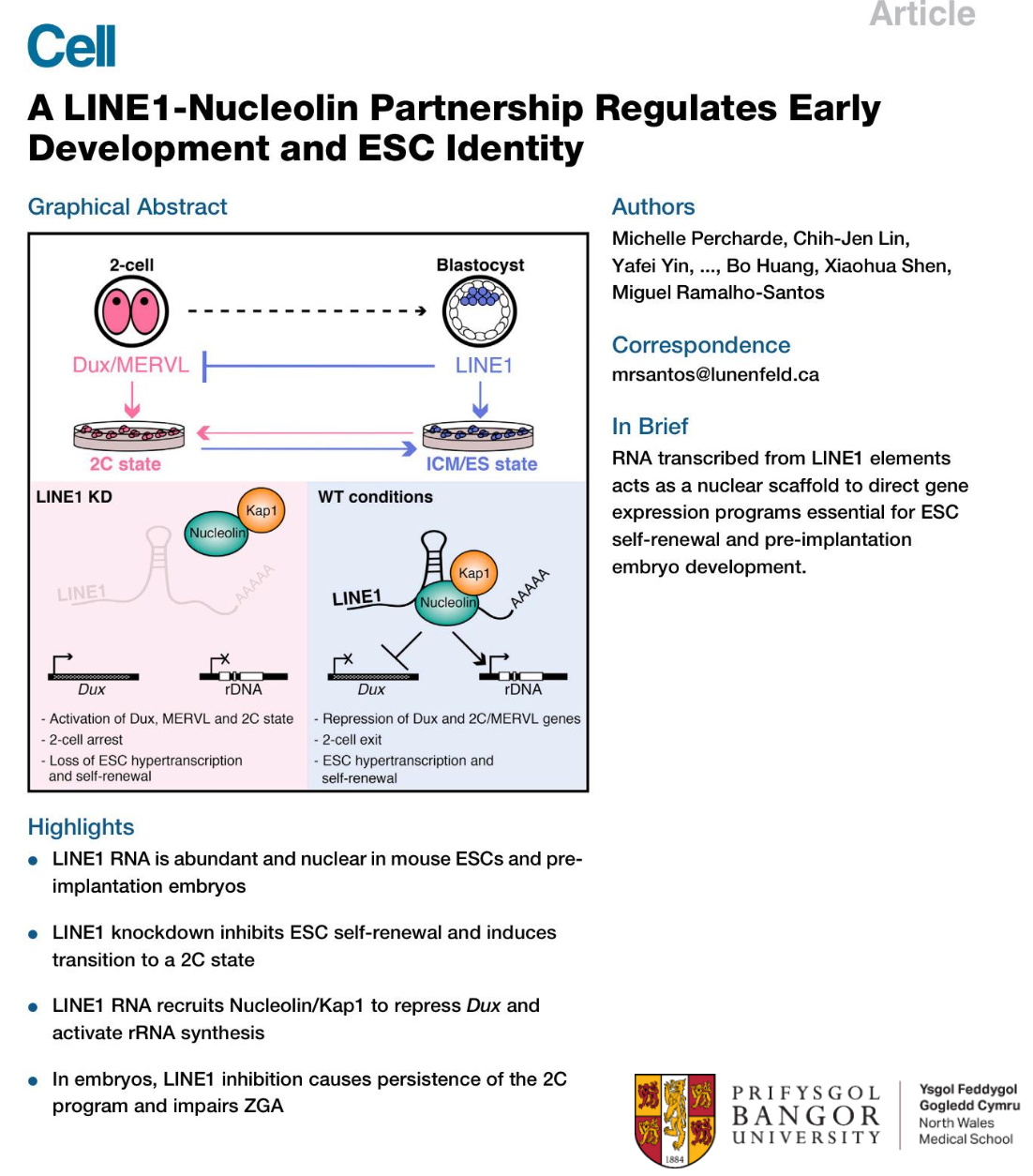
What is a pathogenic role of transposons or TE-encoded transcripts in embryonic & somatic cells?
Example of a pathogenic role: Haemophilia
Pathogenic LINE1 insertions in the coagulation factor VIII gene.
However, transposon or transcribed TEs induced ‘disease’, due to direct ‘transposition’ of the element into a new location within the genome, is rare.
A nucleosome is the molecular unit of…
Chromatin
What shape is a nucleosome and what parts make it up?
It has a histone octamer:
Central H3-H4 tetramer
Flanked by two H2A-H2B dimers
[alpha-helix - loop - alpha-helix] motif
A Linker histone H1 binds to the nucleosome core around the DNA entry and exit points
![<p>It has a histone octamer:<br>Central H3-H4 tetramer <br>Flanked by two H2A-H2B dimers <br>[alpha-helix - loop - alpha-helix] motif<br>A Linker histone H1 binds to the nucleosome core around the DNA entry and exit points </p>](https://knowt-user-attachments.s3.amazonaws.com/d723f26e-0adb-45a2-a45a-254a7bf88d97.png)
How many base pairs of DNA in the histone octamer of a nucleosome and how is it arranged?
147bp of DNA wrapped in 1.65 left hand turns around the octamer. The DNA double helix sharply bends every ~10bp, when the DNA major groove faces towards the nucleosome core. Arginine stabilises these bends.
Name 4 post-translational modifications involved in epigenetics:
Acetylation
Methylation
Phosphorylation
Ubiquitination
what is higher order folding?
the process where chromatin is folded and compacted to form chromatin fibres and eventually mitotic chromosomes
what length do chromatin fibres get folded into?
10nm - beads on a string - nucleosomes linked by 20-80bp of linker DNA
THEN
30nm:
solenoid: packed into a helix with bent linkers
zigzag: connected by straight linkers with alternative stacking
what is the role of H1 in higher order folding of chromatin into fibres?
it stabilises linker DNA orientation
what are the differences between heterochromatin and euchromatin?
euchromatin is transcriptionally active as its more opened up so transcriptional machinery have room to get in and attach. Same principle applies for why heterochromatin is transcriptionally inactive.
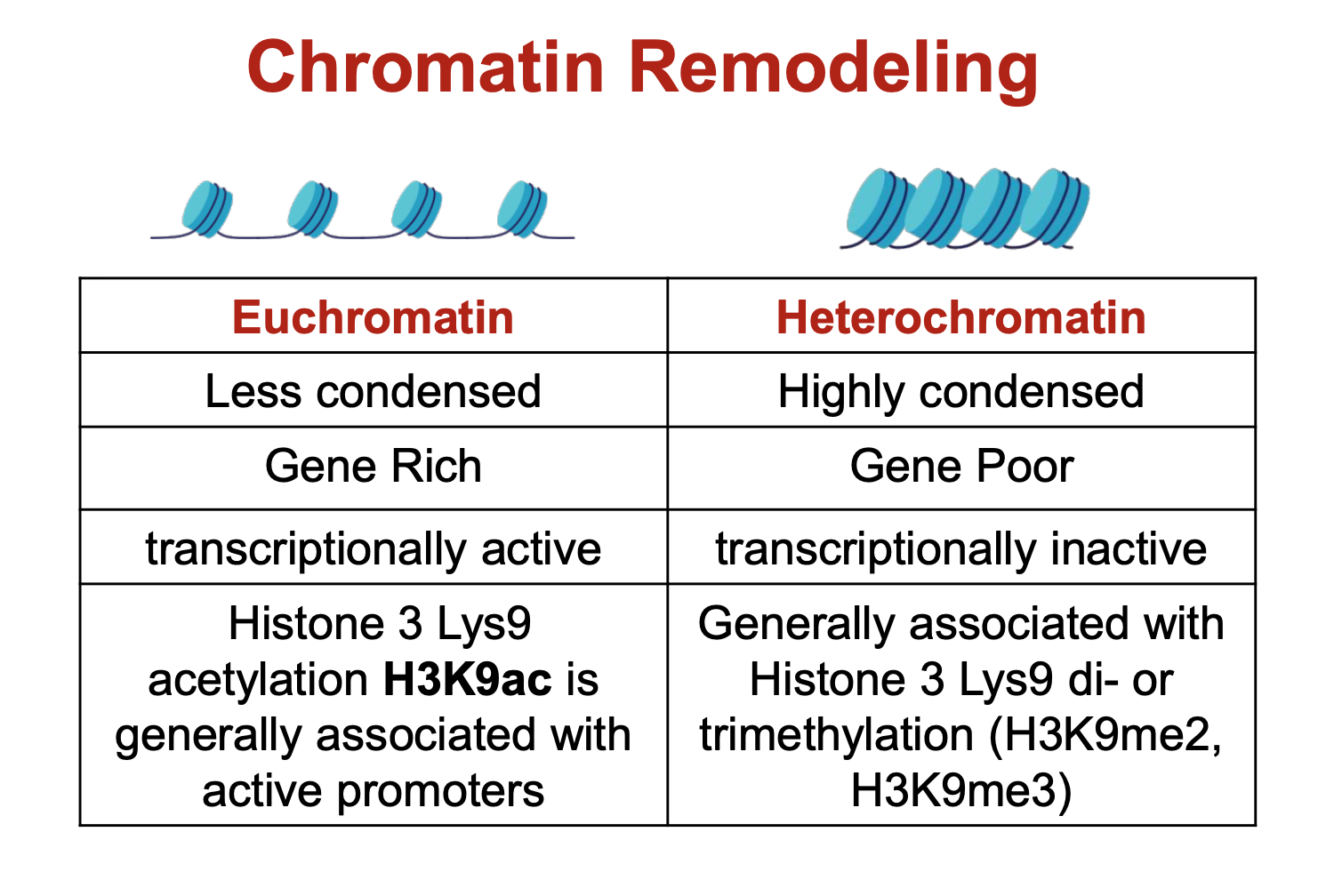
in mitotic chromosome condensation, loop domains are folded into…
loop-scaffolded rosettes
What do Condensing complexes (SMC2/SMC4 & kelvin subunits) do?
Introduce supercoils and axial compression
Maintain sister chromatid until anaphase
What histone modifications are involved in mitotic chromosome condensation?
H3Ser10 - phosphorylation
Global - deacetylation
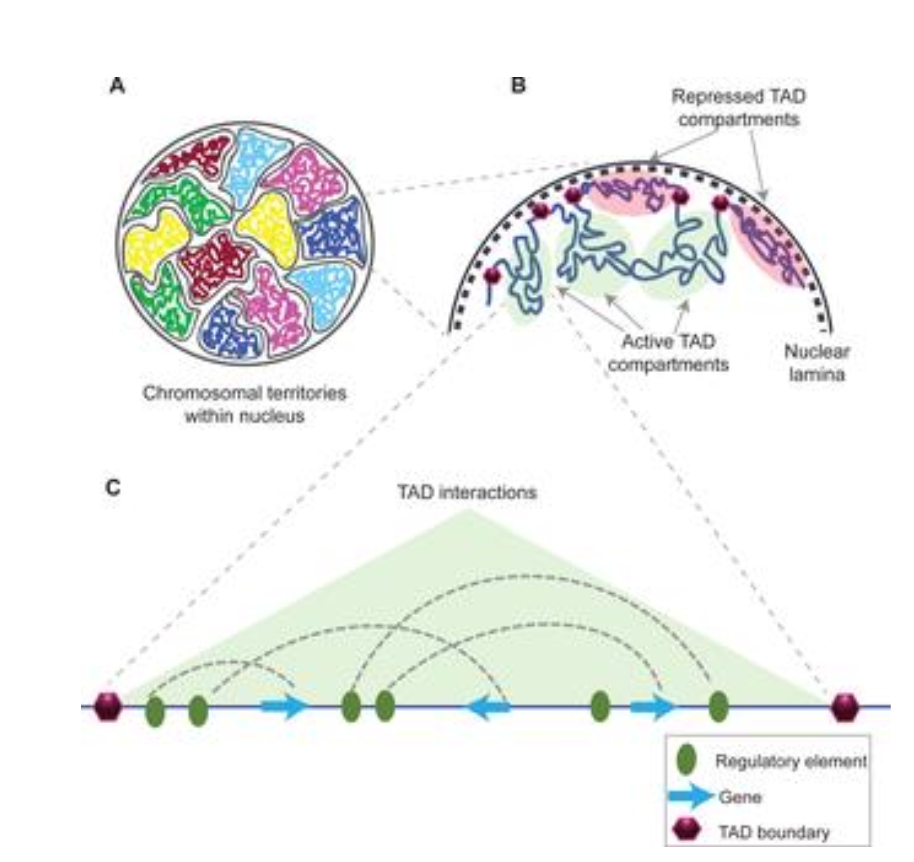
What is Nuclear Architecture?
The special organisation of the genome in different levels of 3D organisation
What are the different levels of 3D organisation in nuclear architecture?
Chromosome territories
Compartments
Topologically Associated Domains (TADs)
Loops (CTCF & Cohesin)
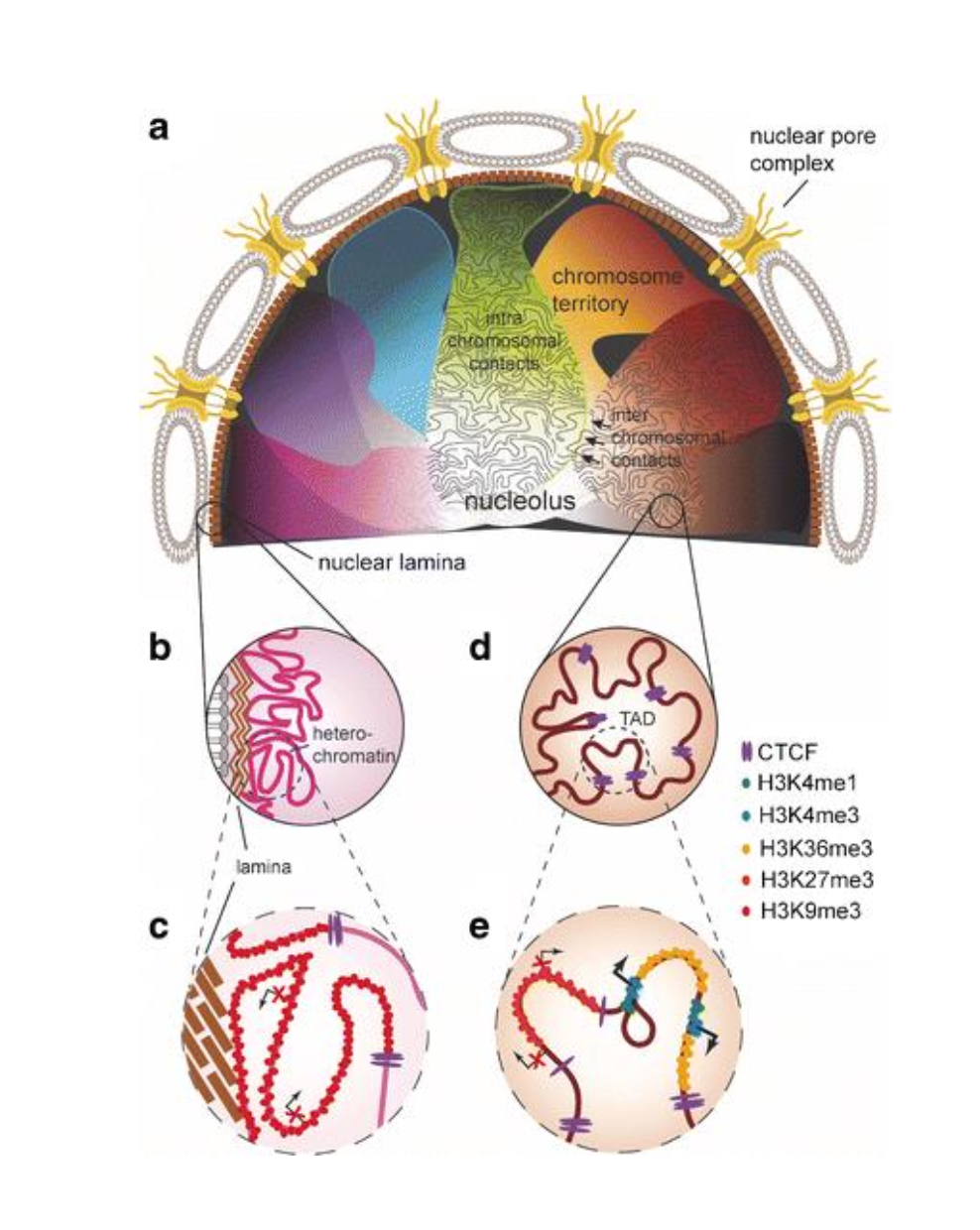
How are the levels of 3D organisation in nuclear architecture identified?
Through experimental approaches:
Microscopy
3C-based methods
Tell me more about different techniques used in Microscopy to detect 3D organisation of nuclear architecture?
Detects transcriptional activity and link spatial organisation to chromatin state.
Uses different techniques such as:
Fluorescence In Situ Hybridisation (FISH) - Fluorescent probes hybridise to specific DNA sequences. It detects chromosome territories, locus radial positioning.
Super-Resolution Microscopy (STORM, SIM, PALM) - Resolves structures below diffraction limit (~20-50 nm). It visualises individual chromatin loops, transcription factories, nucleosome clusters.
Live-Cell Imaging with Fluorescent Tags (e.g. dCas9-GFP) - Tracks dynamic repositioning of loci over time.
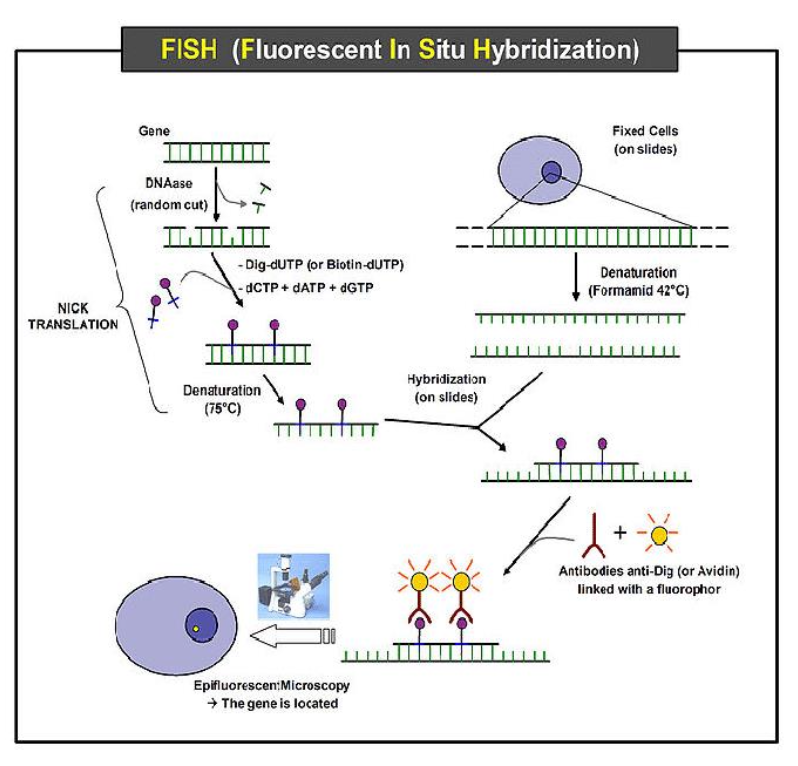
Tell me more about using 3C-based methods in detecting 3D organisation in nuclear architecture?
Two methods:
3C - Chromosome Conformation Capture:
Crosslink DNA-protein complexes with formaldehyde
Restriction enzyme digestion
Ligation of physically close fragments
PCR detection of specific interactions
Hi-C - Genome-wide 3C:
Incorporates biotin-labeled nucleotides at ligation junctions
Sequencing produces contact frequency maps for entire genome
Reveals compartments (plaid pattern), TADs (squares), and loops (dots)
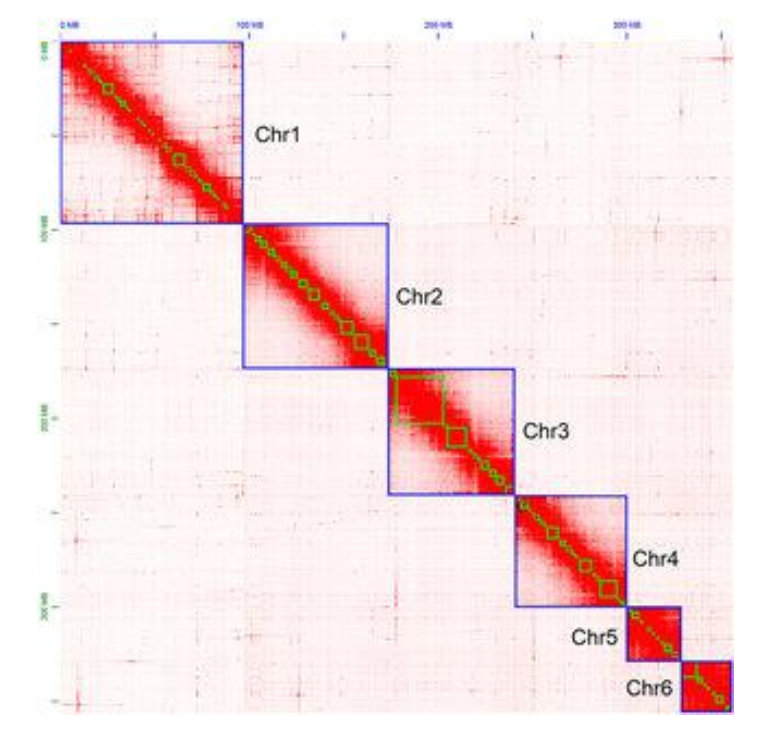
What are Topically Associating Domains (TADs)?
Self-interacting chromatin domains (~100kb-1Mb) that constrain regulatory interactions.
What are the functions of TADs?
Maintains enhancer specificity, preventing miss-expression.
Preserves genome organisation.
What is the boundary molecular composition of TADs?
CTCF:
Zinc-finger DNA-binding protein - binds convergent motifs at TAD borders
Cohesin complex:
Ring-shaped SMC1/SMC3 heterodimer, RAD 21 (klesin), STAG, loaded via NIPBL-MAU2, encircles chromatin.
Other factors:
Control gene promoters, tRNA genes, transcriptionally active loci can stabilise boundaries.
What is the molecular mechanism of the Loop Extrusion Mechanism?
Cohesin loads onto chromatin via NIPBL-MAU2.
ATP hydrolysis drives processive extrusion of DNA through the cohesion ring, enlarging the loop.
Extrusion stops at CTCF-bound sites oriented conversantly leading to a stable loop formation.
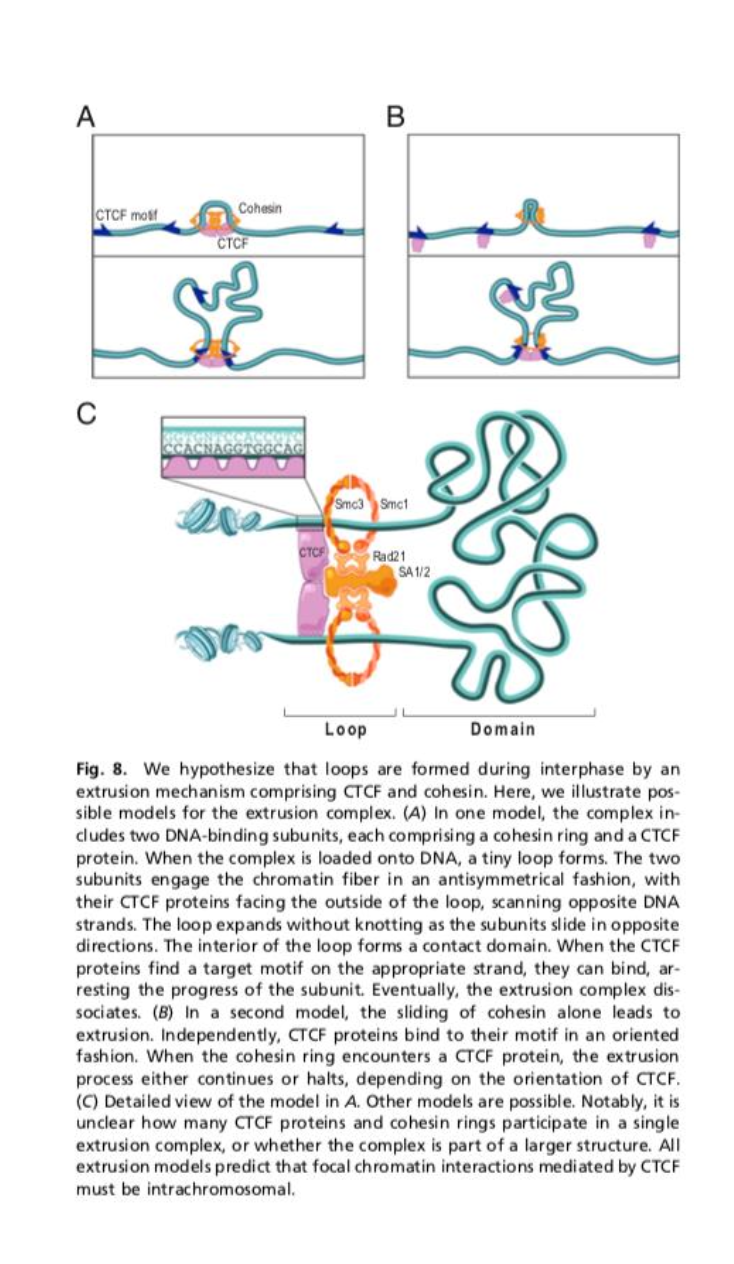
What molecules make up the Loop Extrusion Mechanism?
Cohesin complex:
SMC1/SMC3 heterodimer
RAD 21 (kleisin)
STAG
forms a ring encircling DNA
CTCF:
Binds specific DNA motifs,
acts as a directional barrier
NIPBL-MAU2:
Cohesin loader
ATP dependant.
What provides insulation from neighbouring TADs for the Loop Extrusion Mechanism?
Enhancer-promotor proximity
What is CTCF?
Zinc-finger DNA-binding protein.
Binds convergent motifs at TAD borders
What makes up a Cohesin complex?
SMC1/SMC3 heterodimer
RAD 21 (kleisin)
STAG
complex forms a ring encircling DNA (chromatin)
Why is this loop extrusion mechanism important?
Gene Regulation:
Loops bring enhancers into proximity with promotes (e.g. globin locus)
Insulation:
Prevents ectopic interactions across TAD boundaries
Genome Stability:
Limits inappropriate recombination between domains
Dynamic nature:
Loops/TADs dissolve in mitosis and re-establish in G! phase.