Mol Bio Exam III
1/117
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
118 Terms
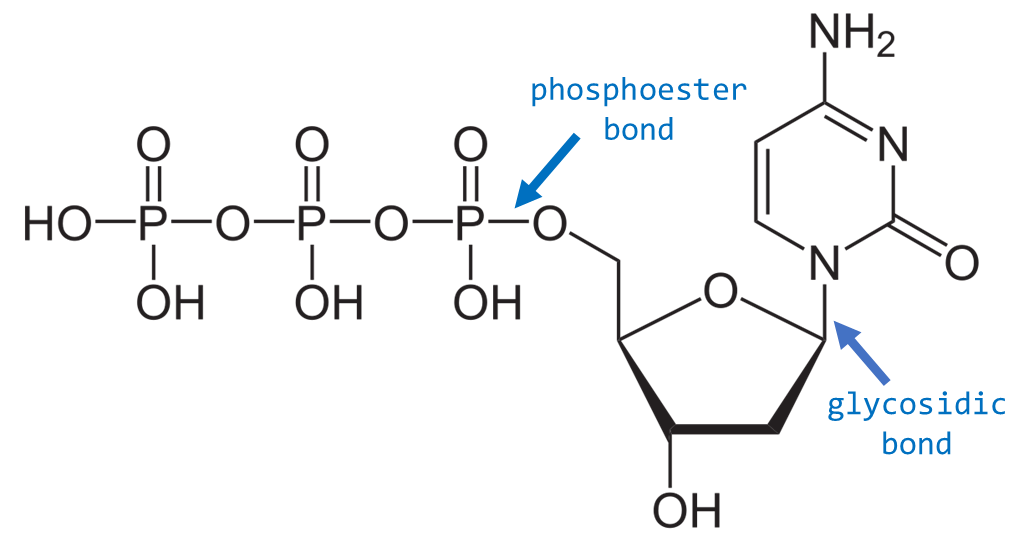
What are the three components of a nucleotide?
1) Deoxyribose sugar (lacks OH at 2' carbon), 2) Phosphate group (attached to 5' carbon), 3) Nitrogenous base (adenine, guanine, cytosine, or thymine)
What is a glycosidic bond?
The bond that attaches the nitrogenous base to the sugar at the 1' carbon position
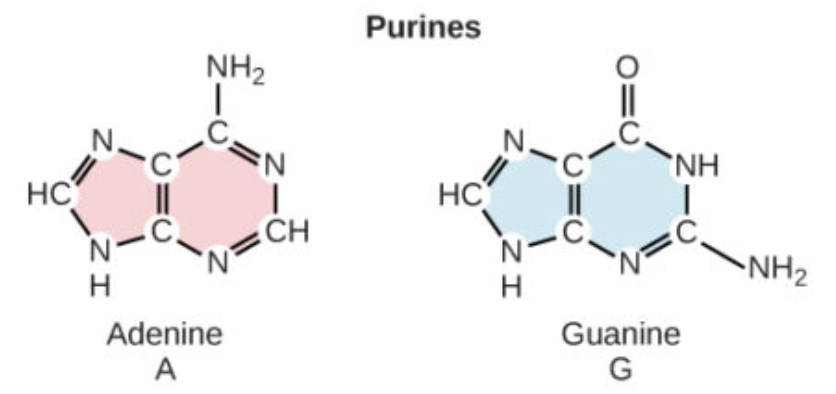
Which bases are purines and how many rings do they have?
Adenine (A) and Guanine (G); they have two rings
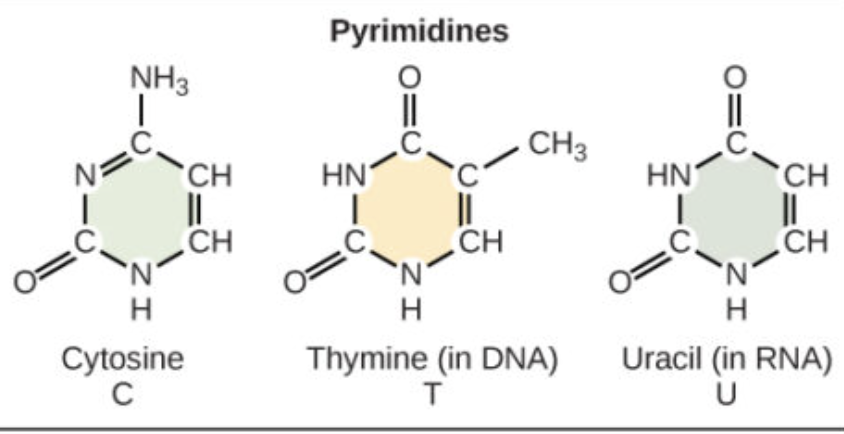
Which bases are pyrimidines and how many rings do they have?
Thymine (T), Cytosine (C), and Uracil (U); they have one ring
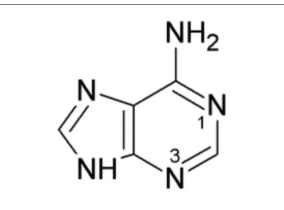
What nitrogenous base is this?
Adenine (Purine)
Unlike G, A has an NH2 rather than a carbonyl at c6
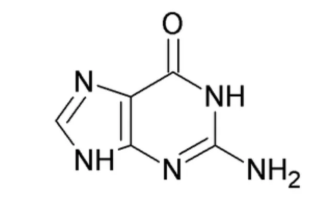
What nitrogenous base is this?
Guanine (Purine)
Unlike A, G has a carbonyl and amine (NH2) group
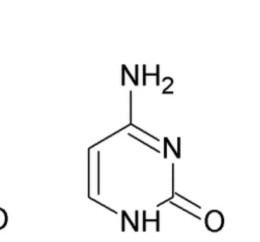
What nitrogenous base is this?
Cytosine (pyrimidine)
Has an NH2 on C6
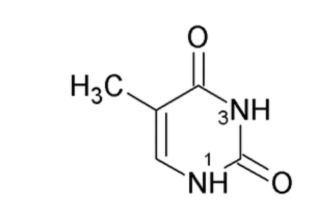
What nitrogenous base is this?
Thymine (Pyrimidine)
Has a methyl group on C5 and a carbonyl on c6
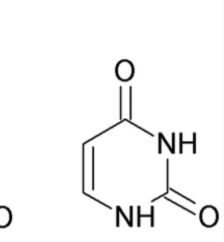
What nitrogenous base is this?
Uracil (Pyrimidine)
Same structure as Thymine but no methyl at c5. Has a carbonyl on C6
What type of bond links nucleotides together in DNA?
Phosphodiester bonds (between the 3' OH of one sugar and 5' phosphate of the next sugar)
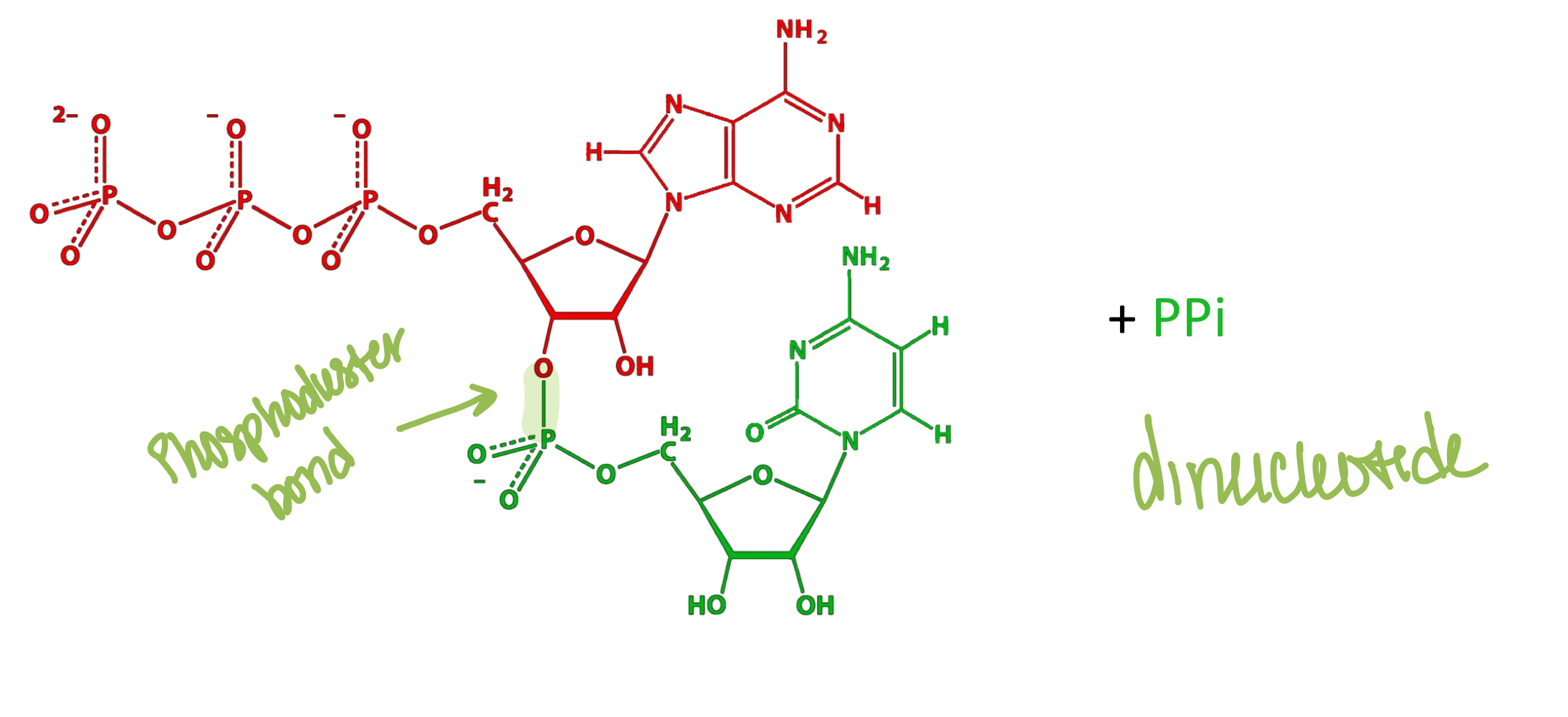
What is the directionality of DNA strands?
DNA strands run 5' to 3', and in double-stranded DNA, the two strands are antiparallel (run in opposite directions)
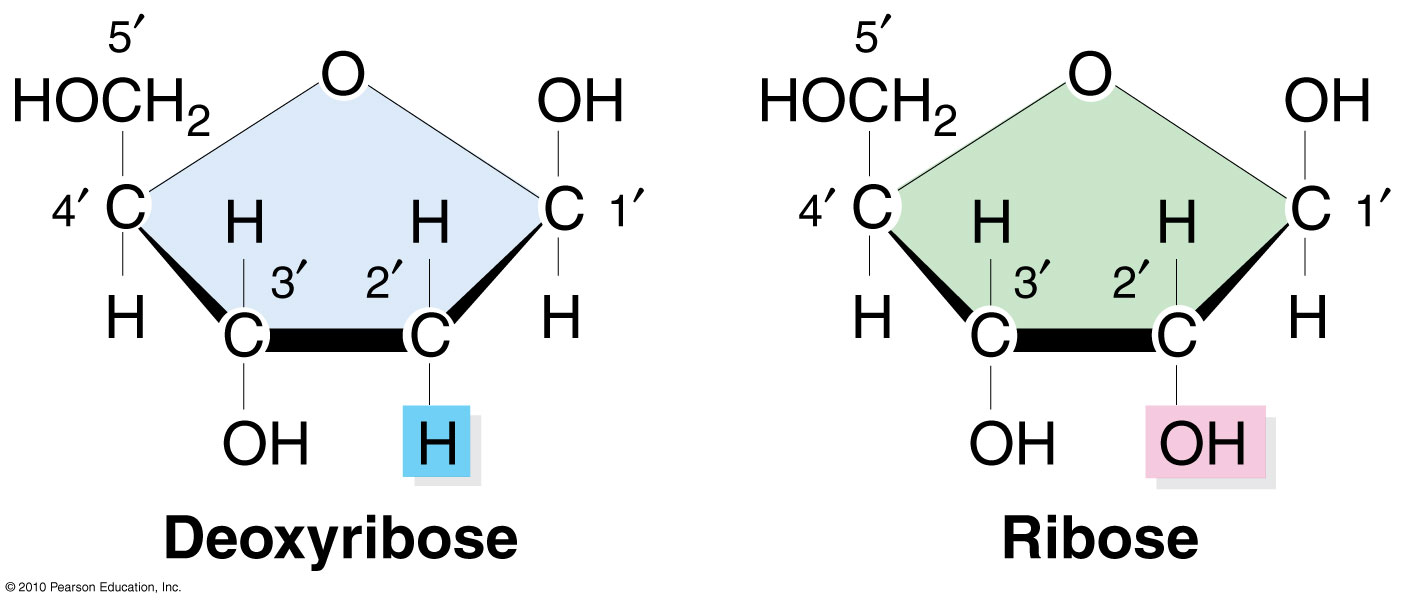
What is the key structural difference between DNA and RNA?
RNA has a hydroxyl group (OH) at the 2' carbon of the ribose sugar, while DNA lacks this OH (deoxyribose)
How many hydrogen bonds form between A-T base pairs?
2 hydrogen bonds
How many hydrogen bonds form between G-C base pairs?
3 hydrogen bonds
What forces stabilize the DNA double helix?
Hydrogen bonds between base pairs and van der Waals forces (base stacking interactions)
What is the functional difference between the major and minor grooves?
The major groove exposes more base identity information, so more proteins bind and base-pairing occurs; the minor groove is narrower with less accessible base information
Why is the major groove important for protein binding?
It exposes more of the bases' functional groups, allowing proteins to "read" the DNA sequence and for base-pairing to occur
What is the first layer of fidelity in DNA replication?
Correct dNTP selection - the correct dNTP binds stably in the DNA polymerase active site, while incorrect dNTPs quickly unbind
How does DNA polymerase recognize correct base pairs in Layer 1?
Through hydrogen bonding interactions with the minor groove; correct base pairs have H-bond donors/acceptors at similar positions that stabilize binding
What is the second layer of fidelity in DNA replication?
Proofreading by the 3' to 5' exonuclease domain of DNA polymerase (editing site)
What happens during proofreading if an incorrect nucleotide is added?
The incorrect nucleotide unbinds from the active site, and the exonuclease domain cleaves the sugar-phosphate backbone to remove it
What is the third layer of fidelity in DNA replication?
DNA repair processes that occur after replication
What is the overall error rate after all three layers of fidelity?
Approximately 1 error in 1 billion nucleotides (1 in 10^9)
Why can't DNA polymerase start synthesis de novo?
DNA polymerase can only add nucleotides to the 3' end of an existing nucleic acid strand (requires a primer)
What enzyme synthesizes the RNA primer for DNA replication?
Primase (also called DNA Pol alpha)
What is the end replication problem?
At linear chromosome ends, when RNA primers are removed, there's a 3' overhang that cannot be filled in, leading to progressive shortening with each replication
How does telomerase solve the end replication problem?
Telomerase uses its internal RNA template to add repetitive DNA sequences to the 3' overhang, extending it so DNA polymerase can fill in the complementary strand
Why don't somatic cells typically express telomerase?
To limit cell division and prevent uncontrolled growth; constantly long telomeres would lead to perpetual replication and potentially cancer
What characterizes the leading strand?
Template runs 3' to 5' (toward the fork); new strand synthesized continuously; only one primer needed
What characterizes the lagging strand?
Template runs 5' to 3' (away from the fork); new strand synthesized discontinuously as Okazaki fragments; multiple primers needed; requires DNA ligase to join fragments
What is helicase's function in DNA replication?
Separates (unwinds) the two template DNA strands at the replication fork
What is the function of ssBPs (single-strand binding proteins)?
Bind to and stabilize single-stranded DNA on the lagging strand template to prevent re-annealing
What is topoisomerase's function?
Cuts DNA backbones to relieve overwinding/supercoiling stress caused by helicase unwinding
What is the function of the PCNA clamp?
Holds the leading and lagging strand DNA polymerases onto the DNA template for processivity
What is DNA polymerase alpha's role?
Synthesizes the RNA primer (primase activity) at the start of replication
What is DNA polymerase delta's role?
Main polymerase for lagging strand synthesis (synthesizes Okazaki fragments)
What is DNA polymerase epsilon's role?
Main polymerase for leading strand synthesis
What is RNase's function in replication?
Removes RNA primers so they can be replaced with DNA
What is DNA ligase's function?
Seals nicks in the sugar-phosphate backbone, particularly joining Okazaki fragments on the lagging strand
What happens to nucleosomes during replication?
H3-H4 tetramers stay associated with DNA; H2A-H2B dimers unbind and then rebind after the replication fork passes
When are histones synthesized for new nucleosomes?
During G1/S phase transition; histone modifications occur BEFORE the replication fork
What are tautomers?
Rare alternative structural forms of bases that can mispair; they usually change back to normal form before causing problems
What happens if a tautomer isn't removed before DNA pol replicates past it?
A mismatch occurs (single base pair mismatch) that can lead to mutations
What is a silent mutation?
A base substitution that doesn't change the amino acid due to codon redundancy
What is a missense mutation?
A base substitution that changes one amino acid for another in the protein
What is a nonsense mutation?
A base substitution that creates a premature stop codon, truncating the protein
What causes slippery sequence errors?
Sequences with multiple identical repeats where strands can loop during replication, causing insertions or deletions (indels)
What is the trombone model?
A model showing how the leading and lagging strand polymerases work together at the replication fork, with the lagging strand forming a loop
How does the lagging strand polymerase know when to release?
When it bumps into the previous Okazaki fragment, it releases and starts synthesizing a new fragment
What is the difference between proofreading and repair?
Proofreading occurs during DNA replication by DNA polymerase; repair occurs before or after replication by dedicated repair proteins
When does the DNA damage checkpoint occur?
In G1 phase, before DNA replication (S phase)
What types of damage does NER (Nucleotide Excision Repair) fix?
Bulky adducts (large hydrocarbons from carcinogens) and pyrimidine dimers (from UV exposure)
What are the steps of NER?
1) Excision nuclease cuts damaged strand on both sides of lesion, 2) Helicase separates strands, 3) Damaged piece released, 4) DNA pol fills gap, 5) Ligase seals
What is transcription-coupled excision repair?
When RNA Polymerase II detects damage during transcription, pauses, and backs up to allow NER to occur
What types of damage does BER (Base Excision Repair) fix?
Damaged bases from oxidation, hydrolysis, or methylation (NOT mismatches)
What are the key steps in BER?
1) Glycosylase removes damaged base, 2) Endonuclease cuts backbone, 3) DNA pol beta replaces nucleotide(s), 4) Ligase seals
What is an apurinic site?
A DNA site where a purine base has been removed, leaving just the sugar-phosphate backbone
What are the two pathways in BER?
Path A: only damaged base removed and replaced; Path B: "flap" pathway where several bases are replaced
What is an example of damage repaired by BER?
Cytosine deamination to uracil, which is removed by uracil DNA glycosylase
What type of damage does NHEJ repair?
Double-strand breaks from UV radiation and chemical mutagens
Why is NHEJ considered error-prone?
It just ligates cut ends together without a template; if multiple cuts occur, pieces could be rejoined incorrectly; always leaves a small deletion ("scar")
What are the steps of NHEJ?
1) Repair proteins (Ku) bind exposed ends, 2) Remove several nucleotides leaving overhang, 3) DNA pol fills gaps, 4) Ends ligated
What types of errors does MMR (Mismatch Repair) fix?
Incorrectly paired bases and indels in repetitive sequences
When does mismatch repair occur?
Shortly after DNA replication
What are the bacterial mismatch repair proteins and their eukaryotic homologs?
Bacterial: MutS and MutH; Eukaryotic: MSH and MLH (mutations linked to many cancers)
What are the steps of MMR?
1) MSH binds mismatch, 2) MLH cuts nascent strand backbone, 3) Section of nascent strand removed, 4) DNA pol fills gap, 5) Ligase seals nick
What is required for HDR (Homology Directed Repair)?
Homologous DNA sequences (sister chromatids after replication or homologous chromosomes)
What type of damage does HDR repair?
Double-strand breaks
What is the key enzyme in HDR?
Rad51 enzyme, which facilitates strand invasion
What are the steps of HDR?
1) DS break occurs and repair proteins bind 2) 5’ exonuclease degrades the 5’ end 3) Rad51 facilitates strand invasion into homologous sequence, 4) DNA pol extends strand using homologous template, 5) DNA uninvades, 6) Ligase seals
What is gene conversion?
When sequence differences between homologous strands are resolved, potentially changing the sequence to match the template
What are homologous chromosomes?
One maternal and one paternal chromosome that are similar in structure, contain the same genes but possibly different alleles (same chromosome number)
Why is recombination necessary?
Essential for proper chromosome segregation during meiosis I; creates genetic diversity
What initiates homologous recombination?
A double-strand break followed by 5' end resection, creating 3' overhangs
What happens during strand invasion?
The 3' overhang from one chromosome invades and base pairs with the complementary strand of the homologous chromosome
What is a Holliday junction?
The crossed-strand intermediate structure formed during recombination when both strands have invaded
What are the two pathways for resolving Holliday junctions?
Pathway A: Cut external strands - produces NON-recombinant chromosomes; Pathway B: Cut both junctions at internal strands - produces RECOMBINANT chromosomes
Do all Holliday junctions lead to genetic recombination (crossover)?
No - only resolution pathway B leads to crossover and recombinant chromosomes; pathway A results in non-recombinant chromosomes
What indicates that crossover occurred?
When the ends of the chromosomes are different from the starting chromosomes (all four strands now differ from original)
Why doesn't pathway A result in crossover?
One chromosome remains basically unchanged; just because DNA was exchanged doesn't mean crossover occurred
What are the two branches of the immune system?
Innate immunity (immediate, non-specific) and Adaptive immunity (specific, forms memory)
What segments make up the heavy chain antibody genes?
V (variable), D (diversity), and J (joining) segments
What segments make up the light chain antibody genes?
V (variable) and J (joining) segments only (no D segment)
What are RSS (Recombination Signal Sequences)?
Conserved sequences flanking V, D, and J gene segments; consist of heptamer and nonamer sequences with 12 or 23 bp spacers
What is the 12/23 rule?
Only gene segments flanked by different spacer lengths (one 12bp, one 23bp) can recombine; prevents inappropriate recombination
What proteins catalyze VDJ recombination?
RAG1 and RAG2 proteins (form the RAG complex)
What does the RAG complex do?
Recognizes and binds RSS motifs; catalyzes precise double-stranded breaks between RSS and gene segments
What structures are created by RAG cleavage?
Hairpins at the coding ends and blunt ends at the signal ends
What is Artemis's role in VDJ recombination?
An endonuclease activated by DNA-PKcs that cleaves hairpin loops to generate single-stranded overhangs
Why does Artemis add diversity?
The cleavage site is variable (imprecise cutting), creating different overhang lengths
What is TdT (Terminal deoxynucleotidyl Transferase)?
An enzyme that randomly adds non-templated (N) nucleotides to exposed DNA ends before ligation
When does TdT add nucleotides?
After hairpin opening but before ligation
What proteins stabilize and ligate DNA ends in VDJ recombination?
Ku70/80 and XRCC4/Ligase IV
What is a signal joint?
The ligated product of the signal ends (RSS sequences); it has no function and is lost
What creates junctional diversity?
Combination of Artemis's imprecise hairpin cutting and TdT's random nucleotide addition
What is the final product of VDJ recombination?
A rearranged V(D)J exon that codes for the variable region of an antibody or T-cell receptor
How does VDJ recombination create antibody diversity?
Through: 1) Combinatorial diversity (many V, D, J segments), 2) Junctional diversity (imprecise joining), 3) N-nucleotide addition by TdT
Compare DNA polymerase and Telomerase
Similarities: Both add nucleotides to 3' end, both synthesize DNA; Differences: Telomerase has internal RNA template, doesn't require DNA template, adds repetitive sequences, not active in most somatic cells
Compare NER and BER
NER: fixes bulky lesions, removes ~30 nucleotide patch; BER: fixes single damaged bases, removes 1 or few nucleotides
Compare NHEJ and HDR
Both repair double-strand breaks; NHEJ: error-prone, no template needed, works in any cell cycle phase, leaves deletion; HDR: accurate, requires homologous template, works in S/G2 phases