7. Rooting & Polarity Estimation
1/7
Earn XP
Description and Tags
Review this lecture; a lot of it is visual.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
8 Terms
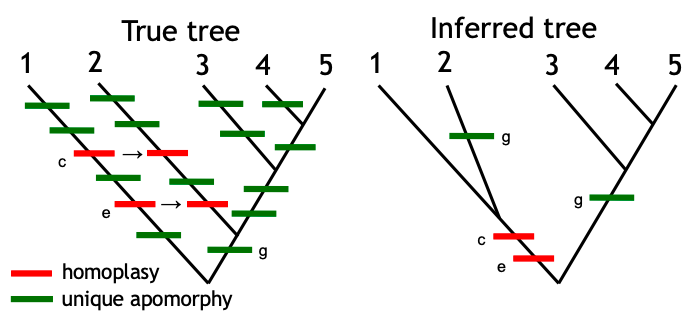
Long Branch Attraction (LBA) occurs when…
When there is a
High frequency of parallel, or
Convergent changes causing sequences (or other characters) to arrive at the same character state
After extended periods or accelerated rates of evolution.
Define ‘Root‘
The earliest node in a tree, the point from which the descendent lineages arose
How to root a tree: Outgroups
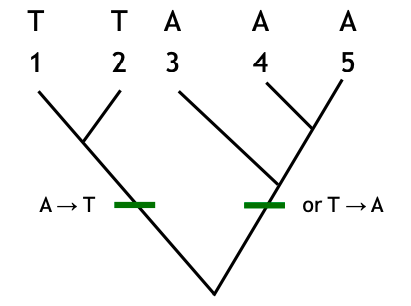
Rooting of trees relates to polarization of character states.
The use of outgroups roots the tree and helps polarize character change.
Ex) Without outgroups: plesiomorphic state could be either A or T.
Multiple outgroups are preferable: Use of multiple outgroups can help resolve these uncertainties, especially if they exhibit the same state for the relevant character.
How to root a tree: The Fossil Record
Can also be used to polarize character change.
Other things being equal, the condition appearing earlier in the fossil record is plesiomorphic.
The sequence of fossil appearance is based on superposition of rock layers.
But: the fossil record is incomplete, and many characters are not preserved in fossils.
How to root a tree: Developmental (embryological) Sequence
Has also been used to polarize character change.
The now-disregarded ”biogenetic law” assumes that the condition appearing earlier in development is plesiomorphic.
“von Baer’s law” is better supported by evidence
How to root a tree: Midpoint Rooting
This method of rooting can be applied to datasets with large numbers of characters, such as DNA sequence data.
Places the root halfway between the two most distant taxa.
Relies on the assumption that evolution is proportional to time (divergence rates are constant).
How to root a tree: Gene-duplication Rooting
This method of rooting can be applied to certain classes of molecular sequence data. It is very compelling when it can be used.
Orthologs are different forms of a single gene that have evolved by speciation within a study group. Functions usually remain similar.
Paralogs are the alternative forms of a duplicated gene that evolve somewhat independently within a study group after the duplication event. Functions usually diverge among paralogs, given time.
Homologs can be either orthologs or paralogs.