Chapter_14_DNA_Structure_Replication_and_Organization
1/66
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
67 Terms
The genetic material of all living organisms is ____.
a. protein
b. deoxyribonucleic acid
c. ribonucleic acid
d. glycoprotein
e. polypeptide
b. deoxyribonucleic acid
Prior to the 1940s, many biologists believed ____, which is composed of 20 different types of ____, was most likely the genetic material.
a. protein; amino acids
b. DNA; amino acids
c. protein; nucleotides
d. DNA; nucleotides
e. RNA; nucleotides
a. protein; amino acids
What happens when living R strain Streptococcus pneumoniae bacteria are mixed with heat-killed S strain Streptococcus pneumoniae bacteria?
a. The S strain bacteria come back to life.
b. The R strain bacteria are killed, and the S strain bacteria remain dead.
c. The R strain bacteria are transformed into S strain bacteria.
d. The S strain bacteria are transformed into R strain bacteria.
e. The R strain bacteria are killed, and the S strain bacteria come back to life.
c. The R strain bacteria are transformed into S strain bacteria.
The alteration of a cell’s hereditary type by the uptake of DNA released by the breakdown of another cell is called ____.
| a. | replication |
| b. | transformation |
| c. | crossing-over |
| d. | excision repair |
| e. | ligation |
b. | transformation |
The transforming principle described by Griffith in his work with Streptococcus pneumoniae is ____.
| a. | a polysaccharide capsule |
| b. | a phospholipid |
| c. | protein |
| d. | RNA |
| e. | DNA |
e. | DNA |
Avery, MacLeod, and McCarty continued Griffith’s work with S. pneumoniae and concluded that DNA was the hereditary material. If protein was the hereditary material instead of DNA, what would happen to mice after injection of heat-killed virulent bacteria mixed with live non-virulent bacteria after treatment with protease?
| a. | The mice would live. |
| b. | The mice would die. |
| c. | The mice would get sick but recover. |
| d. | The mice would grow tumors. |
| e. | The mice would degrade the protein. |
a. | The mice would live. |
In their experiments to identify the transforming principle, Avery, MacLeod, and McCarty used enzymes that break down ____.
| a. | protein, DNA, and RNA |
| b. | protein and RNA only |
| c. | DNA only |
| d. | protein, lipids, and RNA |
| e. | lipids and DNA only |
a. | protein, DNA, and RNA |
In the Hershey and Chase experiment, phage-infected bacteria and viral progeny were radioactive following bacterial infection with 32P-labeled viruses. Why?
| a. | Bacteria had incorporated radioactive proteins into their DNA. |
| b. | Bacteria and viral progeny had incorporated 32P into their DNA. |
| c. | Viral progeny had used the 32P to build new viral proteins. |
| d. | Bacteria had incorporated radioactive proteins into their cell membranes. |
| e. | 32P was left outside the cell during viral infection. |
b. | Bacteria and viral progeny had incorporated 32P into their DNA. |
When Hershey and Chase labeled viruses with radioactive phosphorus, they concluded that the virus injects DNA and not protein into its host because ____.
| a. | most of the radioactive protein was inside the bacteria |
| b. | most of the radioactive DNA was inside the bacteria |
| c. | both radioactive DNA and protein were inside the bacteria |
| d. | neither radioactive DNA nor protein were inside the bacteria |
| e. | entire viruses were inside the bacteria |
b. | most of the radioactive DNA was inside the bacteria |
The T2 bacteriophages used in the Hershey and Chase experiment contain ____.
| a. | DNA only |
| b. | RNA and protein |
| c. | protein, phospholipid, and DNA |
| d. | DNA and protein |
| e. | protein, phospholipid, DNA, and RNA |
d. | DNA and protein |
In the Hershey and Chase experiment, 32P was used to label ____ and 35S was used to label ____.
| a. | RNA; protein |
| b. | protein; DNA |
| c. | phospholipids; protein |
| d. | protein; phospholipids |
| e. | DNA; protein |
e. | DNA; protein |
Suppose that you performed a version of the Hershey and Chase experiment, this time using 32P-labeled viruses that insert their double stranded DNA into the DNA of the cells that they infect. The viral DNA is then treated as part of the cell's own DNA and is replicated during DNA replication and passed onto daughter cells when the cell divides. You infect a population of cells with the 32P-labeled viruses, and then let the infected cells go through two generations of cell divisions. If you then examine the cells, you should find 32P-labeled DNA in ____ of the cells.
| a. | none |
| b. | about 1/4 |
| c. | about 1/2 |
| d. | about 3/4 |
| e. | all |
c. | about 1/2 |
James Watson, Francis Crick, Rosalind Franklin, and Maurice Wilkins are recognized for their contributions to the discovery of the ____.
| a. | presence of nuclein (DNA) in white blood cells |
| b. | role of DNA in cells |
| c. | chemical components of DNA |
| d. | three-dimensional structure of DNA |
| e. | location of DNA in cells |
d. | three-dimensional structure of DNA |
Nucleic acids are long chains of ____.
| a. | amino acids |
| b. | lipids |
| c. | nucleotides |
| d. | sugars |
| e. | peptides |
| c. | nucleotides |
Each DNA nucleotide is composed of ____.
| a. | a six-carbon sugar, a phosphate group, and one of twenty amino acids |
| b. | a five-carbon sugar, a nitrogenous base, and one of twenty amino acids |
| c. | a five-carbon sugar, a phosphate group, and one of four nitrogenous bases |
| d. | a six-carbon sugar, a nitrogenous base, and one of four amino acids |
| e. | a five-carbon sugar, a phosphate group, and one of four amino acids |
c. | a five-carbon sugar, a phosphate group, and one of four nitrogenous bases |
Adjacent nucleotides on a strand of DNA are connected to each other by a(n) ____.
| a. | hydrophobic interaction |
| b. | phosphodiester bond |
| c. | hydrogen bond |
| d. | peptide bond |
| e. | ionic bond |
b. | phosphodiester bond |
If a stretch of human double stranded DNA contains 47% G and C bases, then ____.
| a. | it contains 47% A and T bases |
| b. | there are more pyrimidines than purines |
| c. | there are more purines than pyrimidines |
| d. | it contains 53% A and T bases |
| e. | there are no genes in this stretch of DNA |
d. | it contains 53% A and T bases |
The genome of an organism contains 14% guanine; therefore, it's genome also contains ____% thymine and ____% cytosine.
| a. | 86; 14 |
| b. | 14; 36 |
| c. | 36; 36 |
| d. | 14; 86 |
| e. | 36; 14 |
e. | 36; 14 |
The genome of an organism contains 30% adenine. Based on this, you would predict that this organism's genome also contains 30% ____.
| a. | thymine |
| b. | cytosine |
| c. | each of cytosine and guanine |
| d. | each of thymine and guanine |
| e. | guanine |
a. | thymine |
Purines and pyrimidines are ____.
| a. | Okazaki fragments |
| b. | pentose-phosphate backbones |
| c. | nucleic acids |
| d. | phosphate bases |
| e. | nitrogenous bases |
e. | nitrogenous bases |
How are purines distinguished from pyrimidines?
| a. | Purines are derived from a pair of fused C-N rings, while pyrimidines are derived from a single C-N ring. |
| b. | Pyrimidines are derived from a pair of fused C-N rings, while purines are derived from a single C-N ring. |
| c. | Purines have a carbon-containing ring, while pyrimidines have a nitrogenous ring. |
| d. | Pyrimidines have a carbon-containing ring, while purines have a nitrogenous ring. |
| e. | Purines are found only in DNA, while pyrimidines are found only in RNA. |
a. | Purines are derived from a pair of fused C-N rings, while pyrimidines are derived from a single C-N ring. |
. In DNA, the purines are ____.
| a. | thymine and cytosine |
| b. | adenine and cytosine |
| c. | adenine and guanine |
| d. | thymine and adenine |
| e. | guanine and thymine |
c. | adenine and guanine |
In DNA, the pyrimidines are ____.
| a. | thymine and cytosine |
| b. | adenine and cytosine |
| c. | adenine and guanine |
| d. | thymine and adenine |
| e. | guanine and thymine |
a. | thymine and cytosine |
Which nucleotide sequence is complementary to the DNA sequence 5'-GACGTT-3'?
| a. | 5'-TCATGG-3' |
| b. | 3'-TCATGG-5' |
| c. | 3'-CTGCAA-5' |
| d. | 3'-AGTACC-5' |
| e. | 5'-TTGCAG-3' |
c. | 3'-CTGCAA-5' |
Wilkins and Franklin studied the structure of DNA using ____.
| a. | molecular scale models of nucleotides |
| b. | X-ray diffraction |
| c. | computer-assisted graphics |
| d. | electron microscopy |
| e. | light microscopy |
b. | X-ray diffraction |
A DNA double helix has two strands that are held together by ____.
| a. | hydrogen bonds |
| b. | ionic bonds |
| c. | hydrophobic interactions |
| d. | phosphodiester bonds |
| e. | covalent bonds |
a. | hydrogen bonds |
The width of a DNA double helix ____.
| a. | is constant |
| b. | is narrower where adenine is present than where cytosine is present |
| c. | is wider where purines are present than where pyrimidines are present |
| d. | varies randomly |
| e. | is wider where pyrimidines are present than where purines are present |
a. | is constant |
The two strands of a DNA double helix are said to be antiparallel. This means that ____.
| a. | the 5' end of one strand is directly paired with the 5' end of the other strand |
| b. | since the double helix twists, it is not perfectly parallel |
| c. | one strand has a negative charge and the other strand has a positive charge |
| d. | the 5' end of one strand is directly paired with the 3' end of the other strand |
| e. | both strands have a negative charge |
d. | the 5' end of one strand is directly paired with the 3' end of the other strand |
The polynucleotide chain of DNA has polarity: the 5¢ end has a bound ____, while the 3¢ end has a bound ____.
| a. | deoxyribose sugar, ribose sugar |
| b. | adenine, thymine |
| c. | cytosine, guanine |
| d. | hydroxyl group, phosphate group |
| e. | phosphate group, hydroxyl group |
e. | phosphate group, hydroxyl group |
Which statement correctly describes DNA base pairing?
| a. | Two hydrogen bonds bind A and T; three hydrogen bonds bind G and C. |
| b. | Two hydrogen bonds bind A and C; three hydrogen bonds bind T and G. |
| c. | Two hydrogen bonds bind G and C; three hydrogen bonds bind A and T. |
| d. | Two hydrogen bonds bind A and T, and two hydrogen bonds bind G and C. |
| e. | Three hydrogen bonds bind A and C, and three hydrogen bonds bind T and G. |
a. | Two hydrogen bonds bind A and T; three hydrogen bonds bind G and C. |
DNA replication is said to be semiconservative because ____.
| a. | the number of nucleotides within genes remains constant |
| b. | half of the DNA in a cell comes from one parent and the other half from the other parent. |
| c. | the same process of DNA replication is used by all organisms |
| d. | the total amount of DNA within an individual remains the same |
| e. | each new DNA molecule is composed of one old strand and one new strand |
e. | each new DNA molecule is composed of one old strand and one new strand |
In the Meselson-Stahl experiment, bacterial DNA was labeled completely with heavy nitrogen (15N) and then grown in the presence of light nitrogen (14N). When only mixed DNA was observed after ONE generation of growth in 14N, what was the conclusion?
| a. | DNA replication is semiconservative |
| b. | DNA replication is conservative |
| c. | DNA replication is dispersive |
| d. | DNA replication is either semiconservative or dispersive |
| e. | DNA replication is either semiconservative or conservative |
d. | DNA replication is either semiconservative or dispersive |
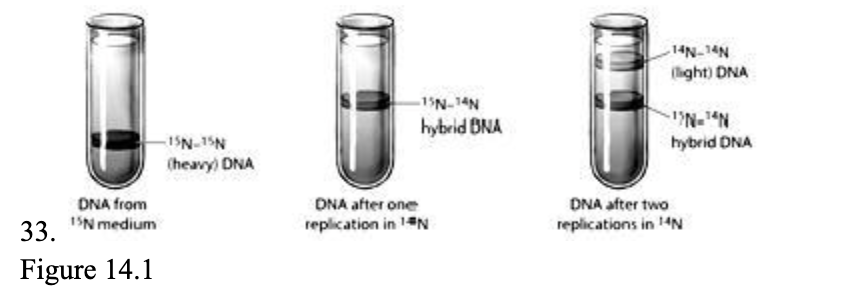
The figure above depicts the result of an experiment to determine how DNA replication occurs. Based on these results, it appears that after replication, each new DNA molecule contains ____.
| a. | either entirely old DNA strands or entirely new DNA strands |
| b. | one old DNA strand and one new DNA strand |
| c. | entirely new DNA |
| d. | some DNA helix regions from old DNA alternating with some DNA regions from new DNA |
| e. | two strands that are each a mix of old and new DNA |
b. | one old DNA strand and one new DNA strand |
DNA polymerase ____.
| a. | synthesizes a short RNA segment to begin DNA replication |
| b. | adds nucleotides to the 5' end of an existing strand to synthesize a new DNA strand |
| c. | adds nucleotides to the 3' end of an existing strand to synthesize a new DNA strand |
| d. | seals nicks between adjacent segments of DNA |
| e. | unwinds the DNA double helix at the origin of replication |
c. | adds nucleotides to the 3' end of an existing strand to synthesize a new DNA strand |
Topoisomerase functions by ____.
| a. | creating cuts in the DNA to relieve over-twisting and strain ahead of the replication fork |
| b. | binding DNA polymerase to hold it tightly to the template DNA |
| c. | unwinding the DNA double helix to expose the template strands for replication |
| d. | binding the newly synthesized DNA to re-twist it into a double helix after replication |
| e. | reading the DNA template and synthesizing a complementary strand of DNA |
a. | creating cuts in the DNA to relieve over-twisting and strain ahead of the replication fork |
The active site of DNA polymerase is similar in ____.
| a. | archaea and bacteria only |
| b. | archaea, bacteria, and eukaryotes |
| c. | archaea and eukaryotes only |
| d. | none of these groups |
| e. | bacteria and eukaryotes only |
b. | archaea, bacteria, and eukaryotes |
DNA ligase closes nicks between ____ by forming ____ bonds.
| a. | nitrogenous bases; hydrogen |
| b. | RNA fragments; noncovalent |
| c. | DNA and RNA fragments; covalent |
| d. | DNA fragments; peptide |
| e. | DNA fragments; covalent |
e. | DNA fragments; covalent |
Helicase ____.
| a. | proofreads the newly synthesized DNA |
| b. | relieves over-twisting ahead of the DNA replication fork |
| c. | rewinds the newly synthesized DNA strands into a double-stranded helix |
| d. | unwinds the double-stranded DNA helix |
| e. | binds DNA polymerase to the template strand |
d. | unwinds the double-stranded DNA helix |
A short RNA chain is synthesized as the first nucleotides in a new DNA strand by the enzyme ____ to provide a ____ for DNA elongation.
| a. | topoisomerase; 3¢ hydroxyl group |
| b. | primase; 3¢ hydroxyl group |
| c. | primase; 5¢ hydroxyl group |
| d. | DNA polymerase; 5¢ phosphate group |
| e. | DNA polymerase; 3¢ hydroxyl group |
b. | primase; 3¢ hydroxyl group |
Why is DNA ligase most active on the lagging strand during DNA replication?
| a. | The lagging strands contain more short DNA segments than the leading strand, which are joined together by DNA ligase. |
| b. | The lagging strand is synthesized more slowly, and DNA ligase speeds up the DNA polymerase. |
| c. | The lagging strand synthesizes DNA in the 3' ® 5' direction. |
| d. | The lagging strand requires DNA ligase to couple the RNA primer to the Okazaki fragments. |
| e. | DNA ligase performs the function of RNA primase on the lagging strand. |
a. | The lagging strands contain more short DNA segments than the leading strand, which are joined together by DNA ligase. |
Adding nucleotides onto a growing DNA strand during DNA replication in cells occurs in ____.
| a. | the 5' ® 3' direction for the leading strand and the 3' ® 5' direction on the lagging strand |
| b. | either the 5' ® 3' direction or the 3' ® 5' direction on both strands, depending on where replication begins |
| c. | the 5' ® 3' direction only |
| d. | the 3' ® 5' direction for the leading strand and the 5' ® 3' direction on the lagging strand |
| e. | the 3' ® 5' direction only |
c. | the 5' ® 3' direction only |
Reiji Okazaki discovered what are now called "Okazaki fragments" produced during DNA replication. These fragments are ____.
| a. | short lengths of new DNA on the leading strand |
| b. | RNA primers on the lagging strand |
| c. | RNA primers on both the lagging and leading strand |
| d. | short lengths of new DNA on the lagging strand |
| e. | RNA primers on the leading strand |
d. | short lengths of new DNA on the lagging strand |
During DNA replication, the ____ strand is assembled in the ____ direction from helix unwinding and is synthesized by ____ replication.
| a. | leading; opposite; continuous |
| b. | lagging; same; discontinuous |
| c. | leading; same; discontinuous |
| d. | lagging; opposite; continuous |
| e. | leading; same; continuous |
e. | leading; same; continuous |
In eukaryotes, the DNA molecule that comprises a chromosome is ____ and has ____ replication origin.
| a. | circular, one |
| b. | circular, more than one |
| c. | linear, more than one |
| d. | linear, one |
| e. | linear, no |
c. | linear, more than one |
The energy to form the new bonds between nucleotides in a growing DNA strand is provided primarily by ____.
| a. | unwinding of the DNA double helix |
| b. | hydrolysis of pyrophosphate |
| c. | breaking hydrogen bonds between base pairs |
| d. | DNA helicase |
| e. | forming hydrogen bonds between base pairs |
b. | hydrolysis of pyrophosphate |
Imagine that a cell contains a genetic mutation in the gene encoding the primase enzyme, rendering it unable to synthesize RNA strands. Assuming that all of the other enzymes directly involved in DNA replication are still functional in these cells, how much of the DNA replication process would you expect to see in these cells?
| a. | The leading strand would be synthesized, but not the lagging strand. |
| b. | No part of the DNA replication process could occur. |
| c. | The DNA helix would be unwound by helicase, but no new strands will be produced. |
| d. | Both the leading and lagging strand would be synthesized, but pieces of discontinuous strands would not be joined together. |
| e. | DNA replication would still proceed completely since RNA strands are not part of the final product of DNA replication. |
c. | The DNA helix would be unwound by helicase, but no new strands will be produced. |
Telomeres are found ____.
| a. | in the middle of chromosomes |
| b. | at replication origins |
| c. | where DNA strands are joined together |
| d. | within genes |
| e. | at the ends of chromosomes |
e. | at the ends of chromosomes |
In humans, telomerase ____.
| a. | adds telomere repeats in some human cells |
| b. | removes telomere repeats in all human cells |
| c. | adds telomere repeats in all human cells |
| d. | removes telomere repeats in some human cells |
| e. | transcribes telomere repeats in some human cells |
a. | adds telomere repeats in some human cells |
During normal DNA replication, part of the sequence at the ends of linear chromosomes is not copied into the new DNA strands because ____.
| a. | DNA ligase cannot join pieces at the end of a chromosome |
| b. | RNA primers at the beginning of a new strand cannot be replaced with DNA |
| c. | those ends are Okazaki fragments that are lost |
| d. | cells do not need the DNA at the ends of chromosomes |
| e. | the ends of chromosomes are made of protein, not DNA |
b. | RNA primers at the beginning of a new strand cannot be replaced with DNA |
Suppose you take a cell from an adult cow and attempt to use it to produce a clone of that cow. If telomerase is not functioning in that cell or in any of the cell’s progeny, what would you expect to happen to your clone?
| a. | The clone will be unable to grow. |
| b. | When the clone matures, it will most likely have cancer. |
| c. | The clone's cells may divide, but after a certain number of generations, cell division will stop. |
| d. | The lack of telomerase should have no effect on the clone. |
| e. | When the clone matures, it will most likely be sterile. |
c. | The clone's cells may divide, but after a certain number of generations, cell division will stop. |
Chromatin consists of ____.
| a. | DNA only |
| b. | DNA, RNA, and protein |
| c. | DNA and RNA |
| d. | RNA and protein |
| e. | DNA and protein |
e. | DNA and protein |
Nucleosomes are best described as ____.
| a. | prokaryotic DNA associated with nonhistone proteins |
| b. | eukaryotic DNA associated with histone proteins |
| c. | prokaryotic DNA associated with histone proteins |
| d. | eukaryotic DNA associated with nonhistone proteins |
| e. | associated histone and nonhistone proteins |
b. | eukaryotic DNA associated with histone proteins |
The nucleosome core particle consists of ____.
| a. | two chromosomes linked together |
| b. | RNA wrapped around an 8-protein histone complex |
| c. | DNA wrapped around an 8-protein histone complex |
| d. | DNA associated with the nuclear envelope |
| e. | DNA replication enzymes associating to form the replisome |
c. | DNA wrapped around an 8-protein histone complex |
During DNA replication, nucleosomes must ____ ahead of the replication fork and ____ after DNA is replicated.
| a. | disassemble; reassemble |
| b. | reassemble; disassemble |
| c. | unwind DNA; disassociate |
| d. | synthesize primers; proofread |
| e. | cut DNA; ligate DNA |
a. | disassemble; reassemble |
When DNA is replicated, ____.
| a. | parental histones are degraded and entirely replaced by new histones |
| b. | DNA is permanently freed from histone binding |
| c. | the histone proteins are also replicated |
| d. | only some of the histone proteins are replicated |
| e. | the number of histones that bind to each DNA molecule is halved |
c. | the histone proteins are also replicated |
In prokaryotes, the DNA molecule that makes up a chromosome is ____ and has ____ replication origin.
| a. | circular; one |
| b. | circular; more than one |
| c. | linear; more than one |
| d. | linear; one |
| e. | linear; no |
a. | circular; one |
Proofreading by ____ corrects errors that occur during DNA replication.
| a. | DNA polymerase |
| b. | primase |
| c. | telomerase |
| d. | DNA ligase |
| e. | helicase |
a. | DNA polymerase |
Without proofreading, the rate of DNA replication errors in bacteria and eukaryotes is as high as one for every ____ nucleotides assembled.
| a. | 10 to 100 |
| b. | 100,000 to 1,000,000 |
| c. | 1,000 to 10,000 |
| d. | 10,000,000 to 100,000,000 |
| e. | 100,000,000 to 1,000,000,000 |
c. | 1,000 to 10,000 |
After DNA repair enzymes remove an incorrect nucleotide resulting from a replication error, ____ is/are needed to complete the repair.
| a. | primase, DNA polymerase, and DNA ligase |
| b. | DNA polymerase |
| c. | DNA polymerase and DNA ligase |
| d. | primase and DNA ligase |
| e. | primase and DNA polymerase |
c. | DNA polymerase and DNA ligase |
Individuals with xeroderma pigmentosum inherit a faulty DNA repair mechanism. As a consequence, ____.
| a. | they are sterile |
| b. | their cells have no proofreading abilities during DNA replication |
| c. | Okazaki fragments produced during DNA replication cannot be joined |
| d. | they easily develop skin cancer when exposed to sunlight |
| e. | their telomeres are shorter than average |
d. | they easily develop skin cancer when exposed to sunlight |
Variability in offspring is largely a result of ____.
| a. | mutations |
| b. | nucleosomes |
| c. | Okazaki fragments |
| d. | DNA repairs |
| e. | RNA primers |
a. | mutations |
Suppose a DNA replication error is not corrected. After two cell divisions, how many of the four daughter cells contain this mutation (assuming that the mistake was never corrected)?
| a. | none |
| b. | one |
| c. | two |
| d. | three |
| e. | four |
e. | four |
Cancer cells ____.
| a. | maintain telomerase function |
| b. | have longer telomeres than normal cells |
| c. | eventually die due to shortening chromosomes |
| d. | have circular chromosomes |
| e. | cannot replicate their DNA |
a. | maintain telomerase function |
What is the function of the sliding clamp in DNA replication?
| a. | unwind DNA ahead of the replication fork |
| b. | anchor DNA polymerase to the template strand |
| c. | remove DNA polymerase from the template strand |
| d. | anchor primase to the template strand |
| e. | bind to single stranded DNA to stabilize it before it is replicated |
b. | anchor DNA polymerase to the template strand |
The structure of the sliding clamp is ____ in bacteria, archaea and eukaryotes, which likely indicates that ____.
| a. | largely variable; the function of the sliding clamp is different in these organisms |
| b. | largely variable; the function of the sliding clamp is similar in bacteria and archaea but different in eukaryotes |
| c. | highly conserved; the sliding clamp is involved in different processes depending on the organism |
| d. | highly conserved; the function of the sliding clamp is similar in these organisms |
| e. | largely variable; the sliding clamp is not essential for DNA replication |
d. | highly conserved; the function of the sliding clamp is similar in these organisms |
Some research has indicated that increased exercise is linked to increased telomere length in renewable tissues, like blood. Assuming that telomerase levels are not increased by exercise, what might explain the increase in telomere length?
| a. | DNA replication occurred more frequently in individuals who exercised. |
| b. | Cell division occurred more rapidly in individuals who exercised. |
| c. | Telomerase activity was higher in individuals who exercised. |
| d. | Telomerase activity was lower in individuals who exercised. |
| e. | Fewer DNA replication errors occurred in individuals who exercised. |
c. | Telomerase activity was higher in individuals who exercised. |
Cancer treatments may specifically target rapidly dividing cells by using drugs or chemicals that ____.
| a. | promote DNA replication only |
| b. | inhibit DNA repair enzymes only |
| c. | inhibit DNA replication only |
| d. | promote DNA replication or inhibit DNA repair enzymes |
| e. | inhibit DNA replication or DNA repair enzymes |
e. | inhibit DNA replication or DNA repair enzymes |