Exam 1: Ch 1,3,4,6
0.0(0)
0.0(0)
Card Sorting
1/115
Earn XP
Description and Tags
FSU cell structure and functi flash cards
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
116 Terms
1
New cards
What are the universal features of life?
1. All cells store hereditary information in the form of double-stranded DNA molecules
2. All cells replicate their hereditary information via template polymerization
3. All cells transcribe portions of their DNA into RNA molecules
4. All cells use proteins as catalysts
5. All cells translate RNA into protein in the same way
6. Each protein is encoded by a specific gene
7. Life requires a continual input of free energy
2
New cards
Transcription
segments of DNA used as template for mRNA synthesis
3
New cards
Translation
RNA used to synthesize proteins
4
New cards
Gene
Segment of DNA corresponding to specific protein, catalytic, regulatory, or structural RNA molecule
5
New cards
What differentiates eukaryotes from prokaryotes?
* DNA-containing membrane-enclosed nucleus
* Cytoplasm
* Cytoskeleton
* Cytoplasm
* Cytoskeleton
6
New cards
What is the function of the endoplasmic reticulum?
Membrane synthesis, synthesis of materials for secretion
7
New cards
What is the function of the Golgi apparatus?
Receives, modifies, and packages ER molecules.
8
New cards
What is the function of lysosomes?
intracellular digestion (inside of cell)
9
New cards
What is the function of peroxisomes?
cell detoxification
10
New cards
What is cytosol?
everything within the cell membrane besides membrane-bounded organelles
11
New cards
How did the first eukaryotic cell rise?
Archaea absorbed a bacteria (proto-mitochondria), forming an endosymbiotic relationship where the bacteria produced ATP for the archaea
* evidence
* mitochondria share many similar traits with bacteria such as division, DNA, ribosomes.
* evidence
* mitochondria share many similar traits with bacteria such as division, DNA, ribosomes.
12
New cards
Which of the following is not true of all cells?
a. They synthesize proteins on the ribosome
b. They replicate their genome via DNA polymerization
c. They transcribe their genetic information by RNA polymerization
d. They use RNA as a template for genomic DNA polymerization.
a. They synthesize proteins on the ribosome
b. They replicate their genome via DNA polymerization
c. They transcribe their genetic information by RNA polymerization
d. They use RNA as a template for genomic DNA polymerization.
d. They use RNA as a template for genomic DNA polymerization.
13
New cards
What would you NOT expect to find in a bacterial cell?
A. Swimming using flagella
B. Having a cell wall around the plasma membrane
C. ATP production in mitochondria
D. Protein production on the ribosome
A. Swimming using flagella
B. Having a cell wall around the plasma membrane
C. ATP production in mitochondria
D. Protein production on the ribosome
C. ATP production in mitochondria
14
New cards
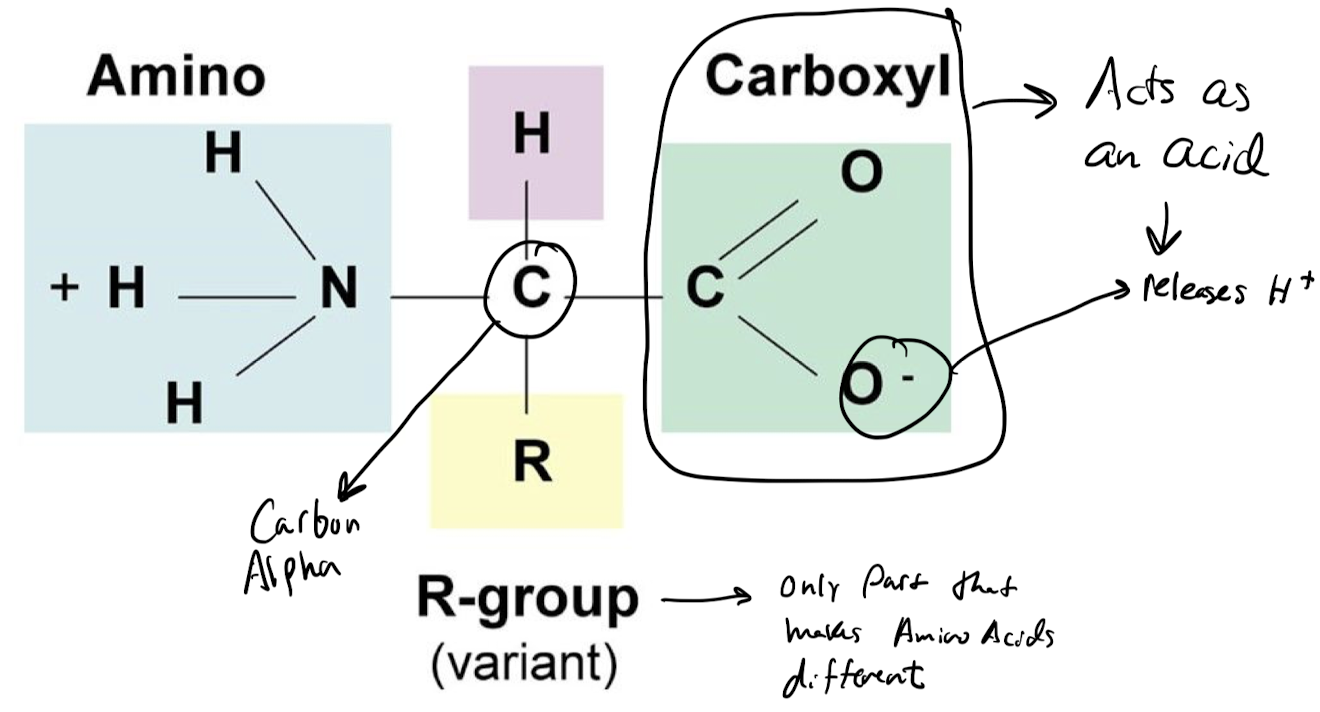
What makes up every protein?
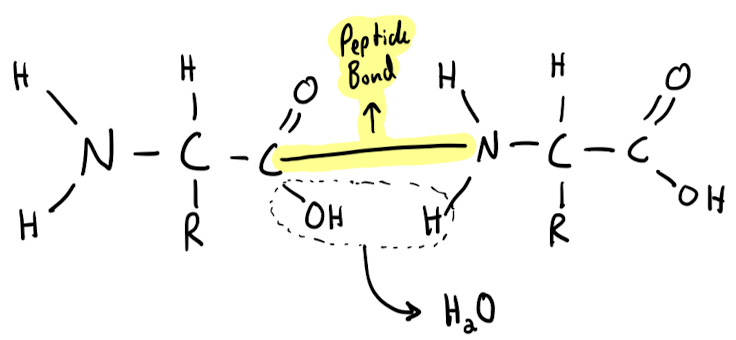
**long unbranched chain** of amino acids, each linked to its neighbor through a **covalent peptide bond**.
15
New cards
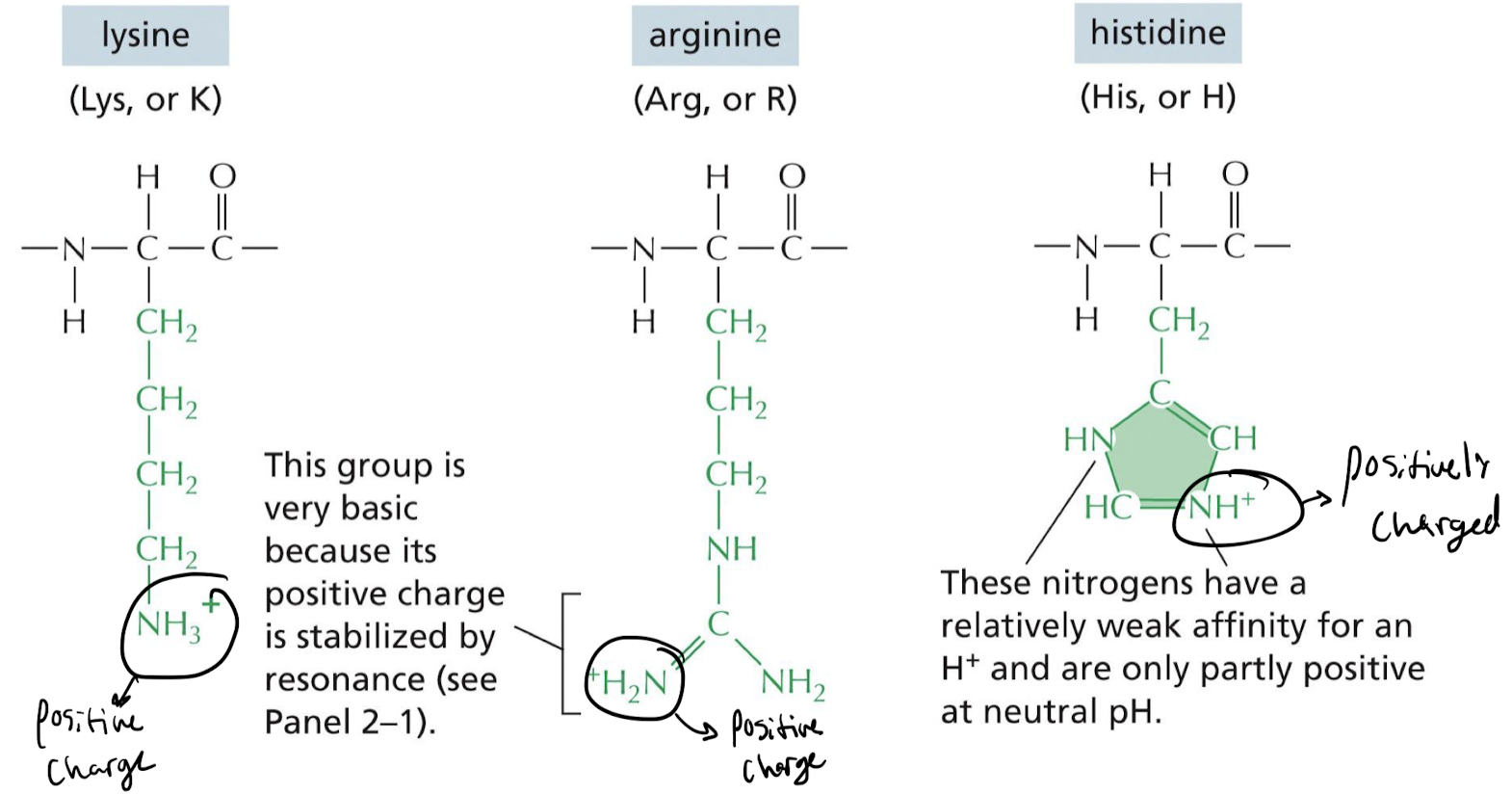
What makes an amino acid side chain basic?
Positively charged N terminal
* ex) NH3+, +H2N, NH+ at N-terminal
* ex) NH3+, +H2N, NH+ at N-terminal
16
New cards
What makes an amino acid side chain acidic?
The amino acid has no charge
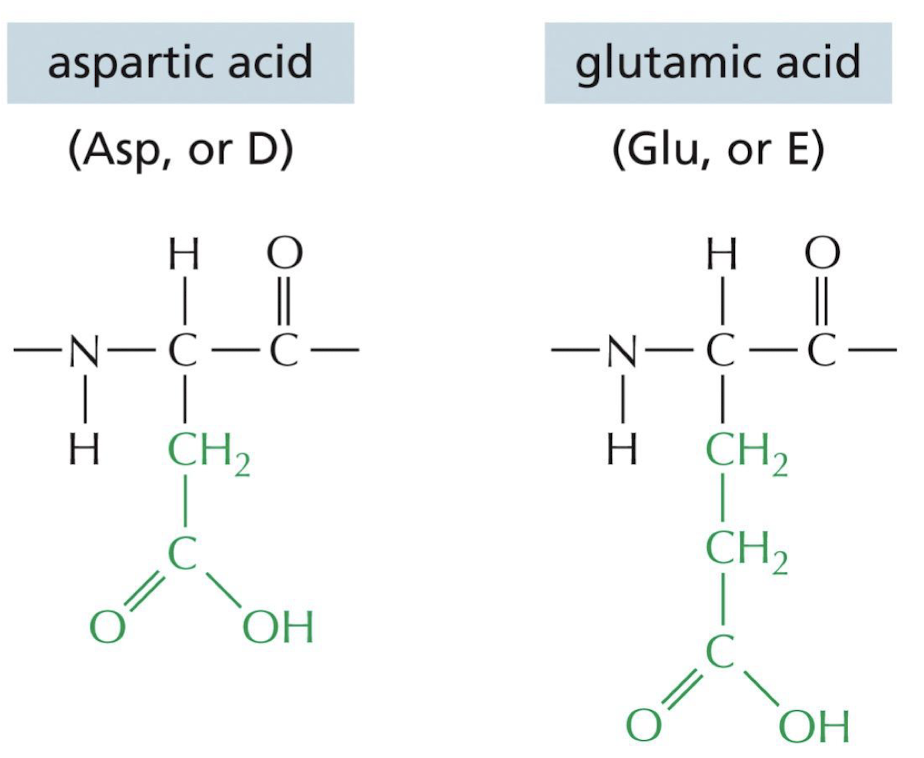
17
New cards
What makes an amino acid side chain uncharged polar?
The amino acid has a polar group attached to them but remain uncharged
* ex) OH or NH2 group attached but still uncharged
* ex) OH or NH2 group attached but still uncharged
18
New cards
What is unique about cystenine?
disulfide bonds can form between two cysteine side chains
19
New cards
What is the primary structure of proteins?
The linear sequence of amino acids
* influences possible bonding angles → impacts overall shape of the protein
* influences possible bonding angles → impacts overall shape of the protein
20
New cards
What is the secondary structure of proteins?
The two patterns that proteins can fold into as a result of hydrogen bonding b/w N-H and C=O groups in **polypeptide backbone**
1. Alpha helix
1. Beta sheet → parallel or antiparallel
1. Alpha helix
1. Beta sheet → parallel or antiparallel
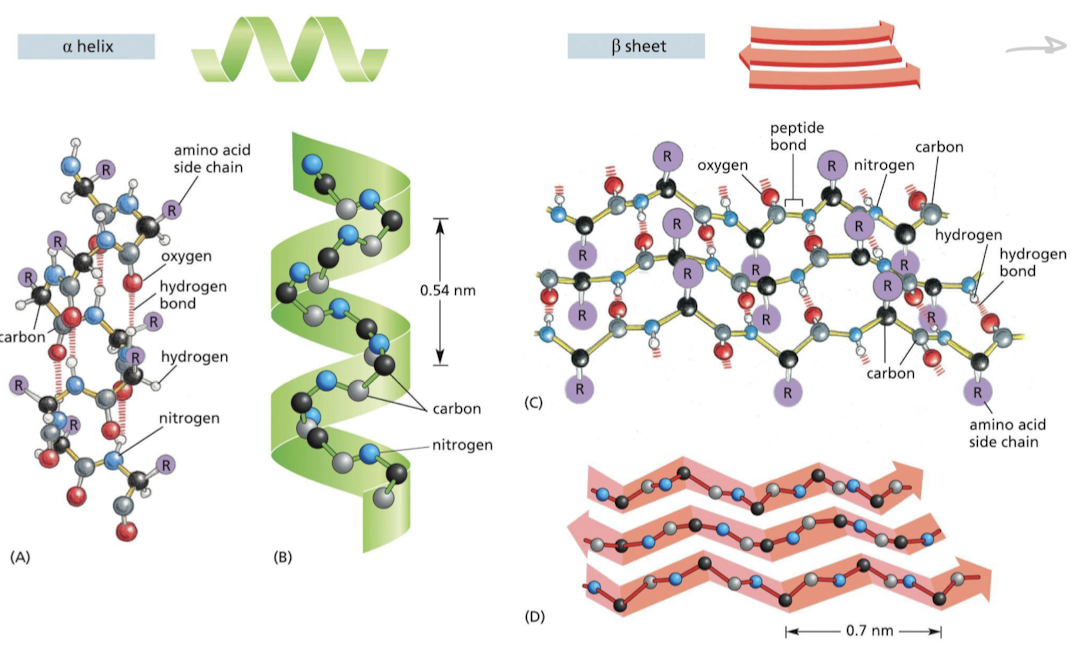
21
New cards
What are the two forms of beta sheets?
Parallel and antiparallel
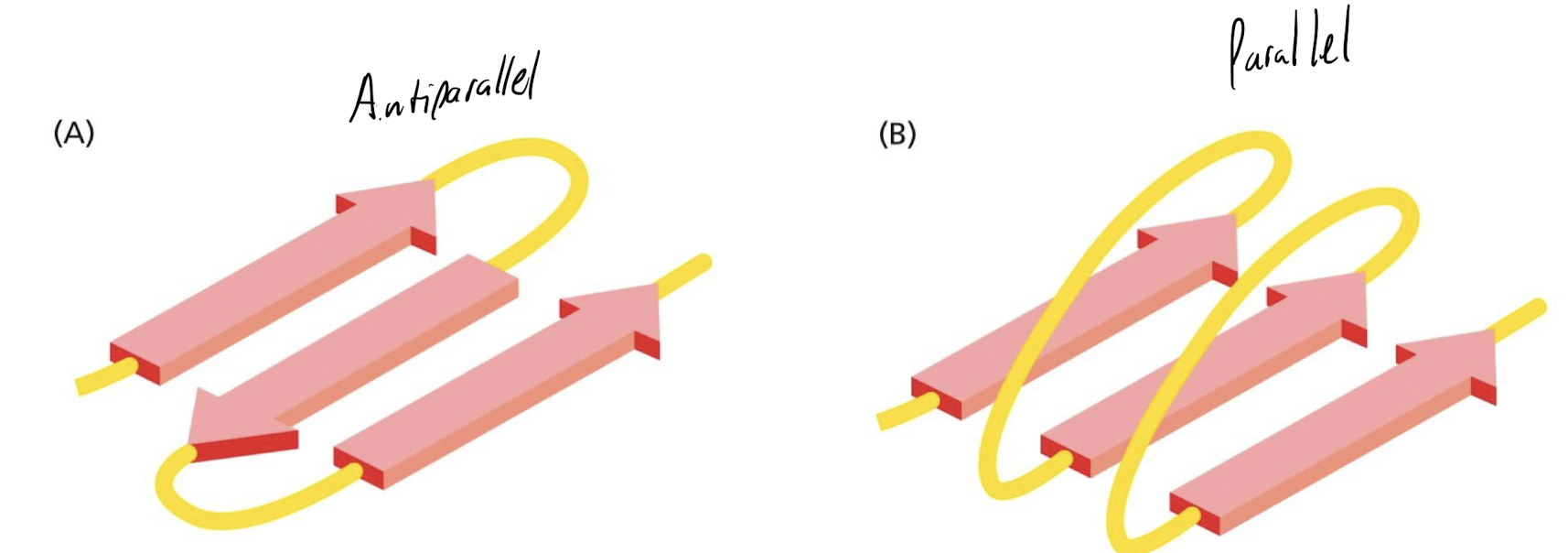
22
New cards
How do alpha helices form?
hydrogen bonding forming between **every fourth peptide bond**
23
New cards
How do coiled-coil structures form?
forms when a helices have most of their non-polar (hydrophobic) side chains on one side.
* ex) a-keratin, myosin
* ex) a-keratin, myosin

24
New cards
What are tertiary structures of proteins?
The full three-dimensional organization of a polypeptide chain -including its alpha helices, beta sheets
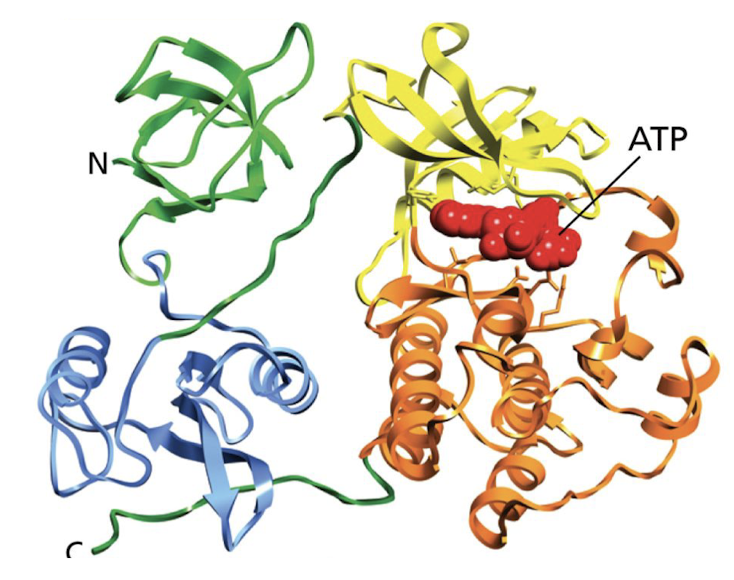
25
New cards
What are quaternary structures of proteins?
protein structures formed by **more than one polypeptide chain**
* ex) hemoglobin
* ex) hemoglobin
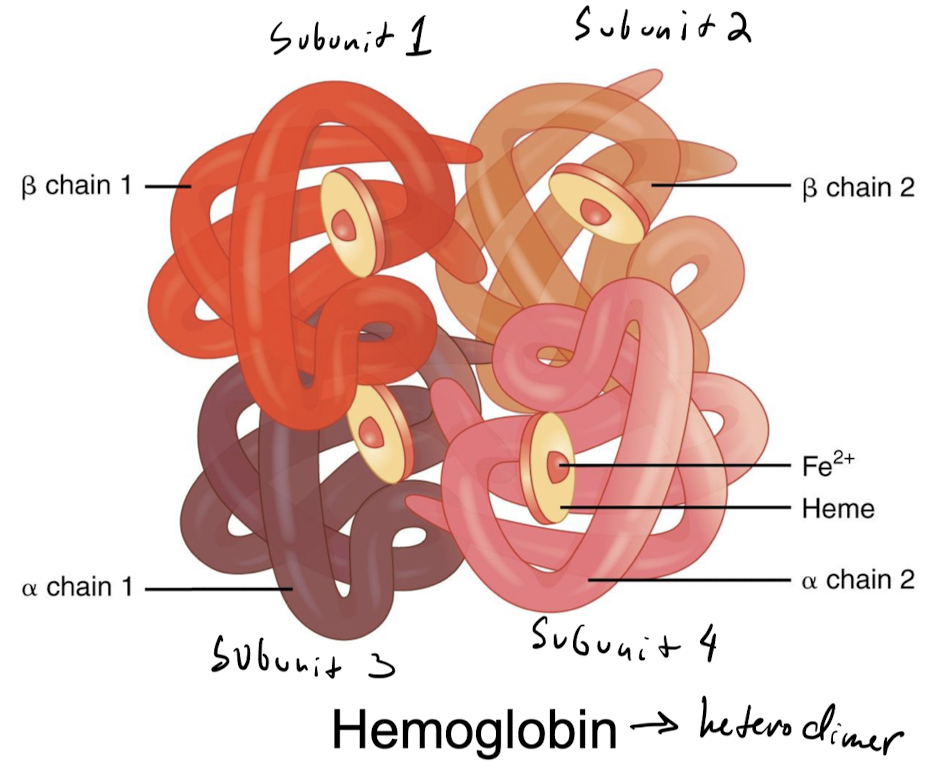
26
New cards
What determines the pattern of folding for proteins?
Weak non-covalent bonds → strong when together
1. hydrogen bonds
2. electrostatic attractions
3. van der Waals attractions
1. hydrogen bonds
2. electrostatic attractions
3. van der Waals attractions
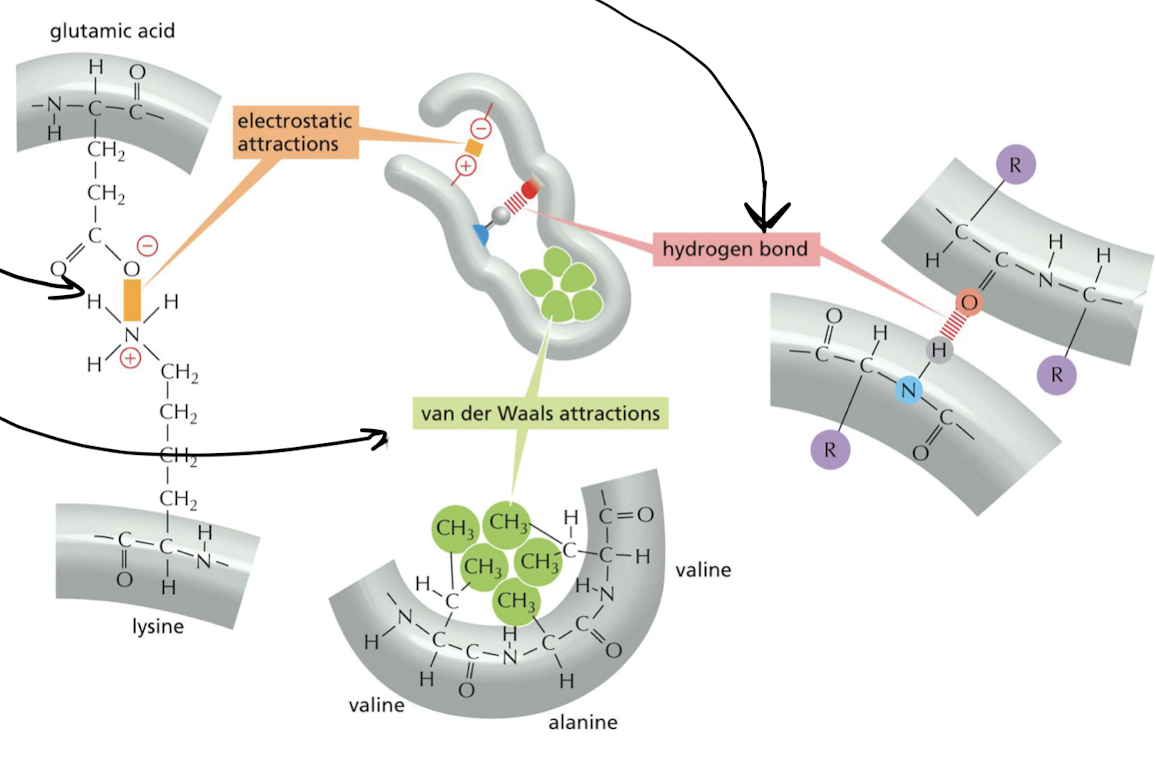
27
New cards
What is the hydrophobic clustering force?
Nonpolar side chains of amino acid tends to be forced together when suspended in H2O
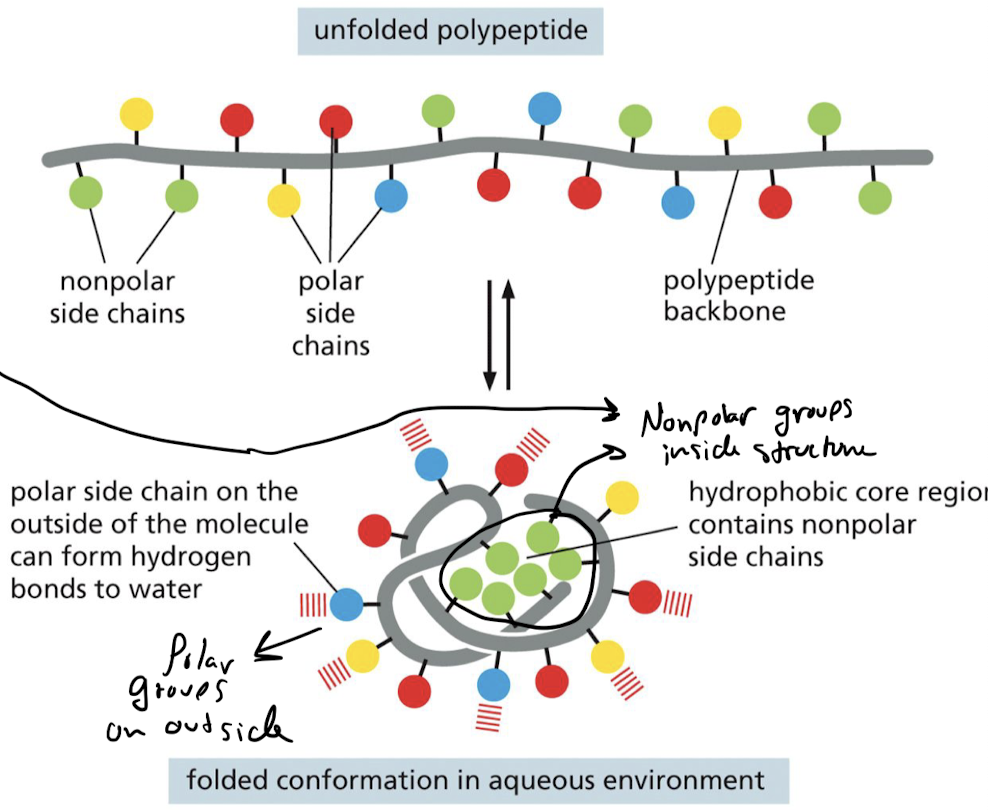
28
New cards
What are covalent cross-linkages and what is their function in protein structure?
* Usually occur on extracellular proteins
* link 2 amino acids of same protein together
* help stabilize the protein
* ex) S-S bond in cysteine
* link 2 amino acids of same protein together
* help stabilize the protein
* ex) S-S bond in cysteine
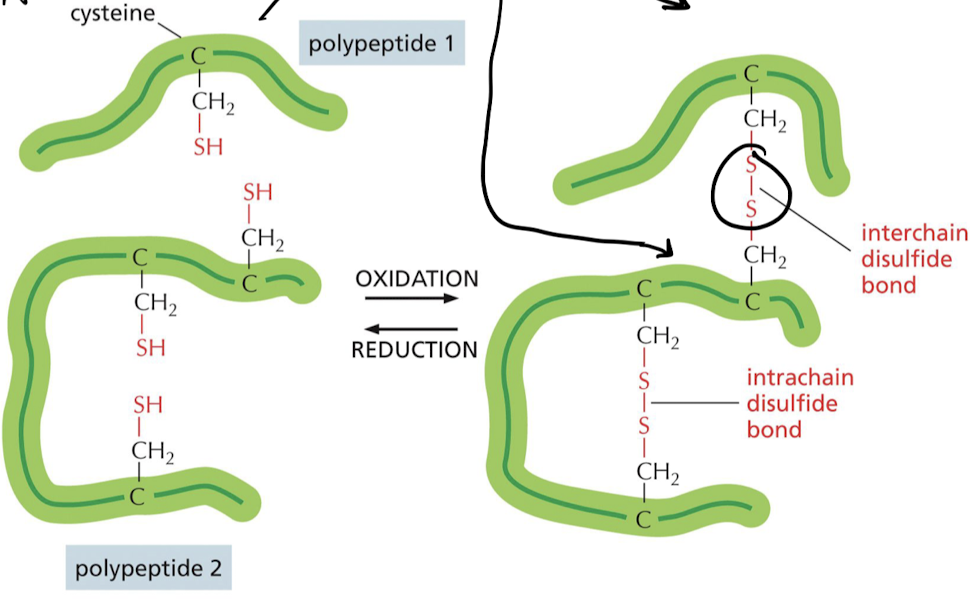
29
New cards
What are protein domains?
sections of one single linear strand that has its own 3D structure, NOT QUATERNARY
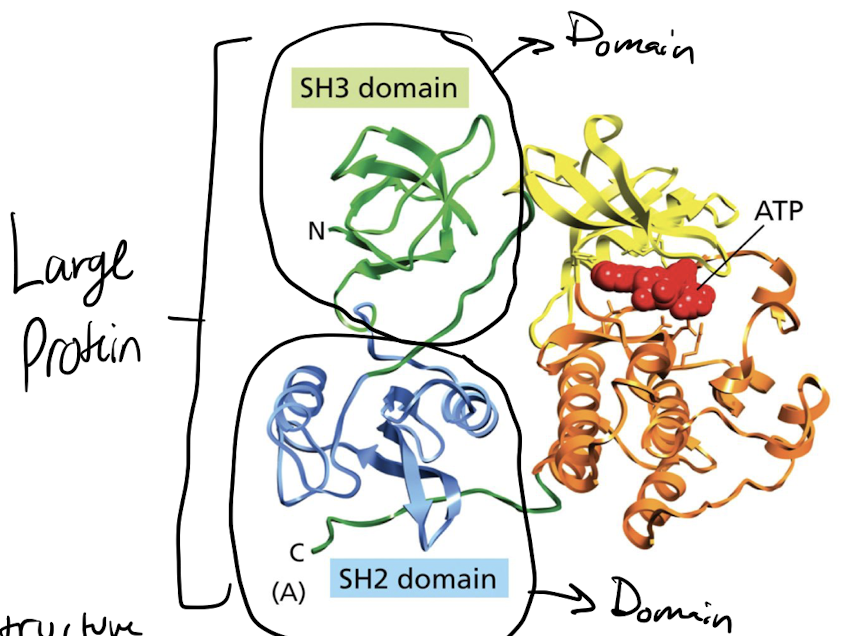
30
New cards
What is domain shuffling?
* Joining of preexisting domains in new combinations.
* Originated from the accidental joining of the DNA sequences that encode each domain, creating a new gene
* Originated from the accidental joining of the DNA sequences that encode each domain, creating a new gene
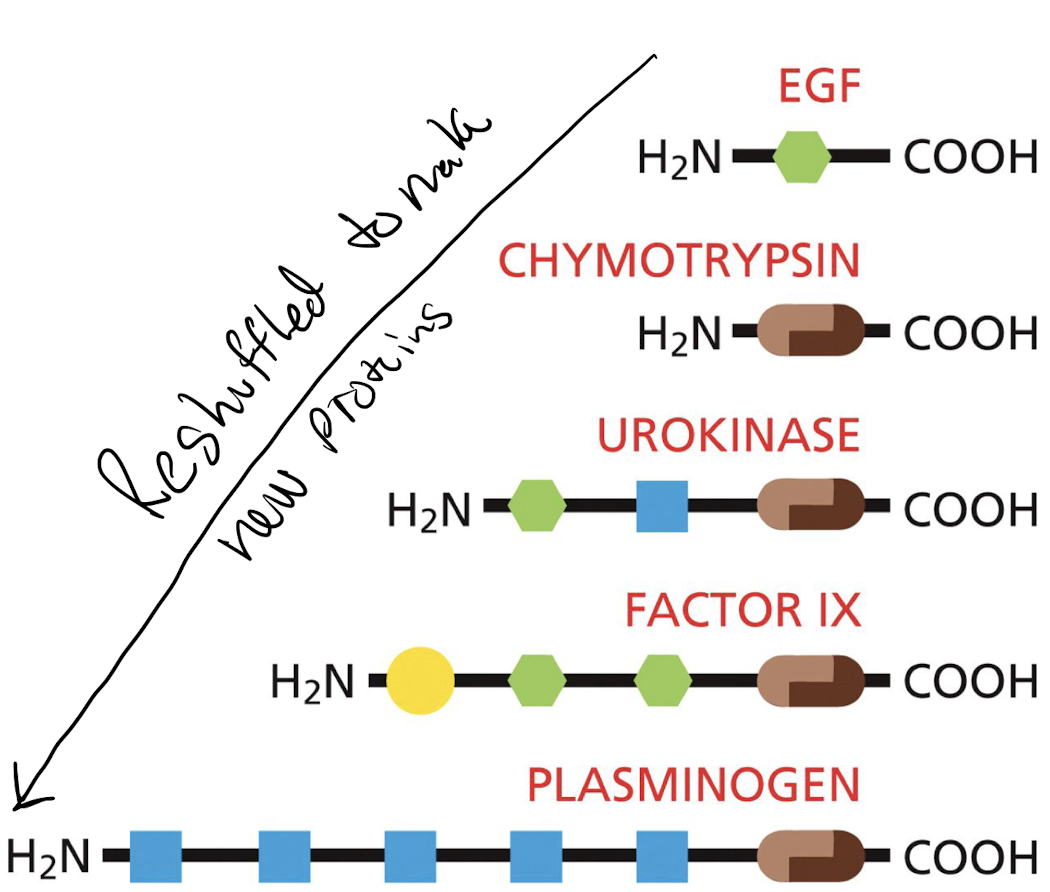
31
New cards
How do protein assemblies form?
have binding site complementary to another region of the surface of the same molecule.
* can only form in a specific way
* ex) Actin forms long helical structure
* can only form in a specific way
* ex) Actin forms long helical structure
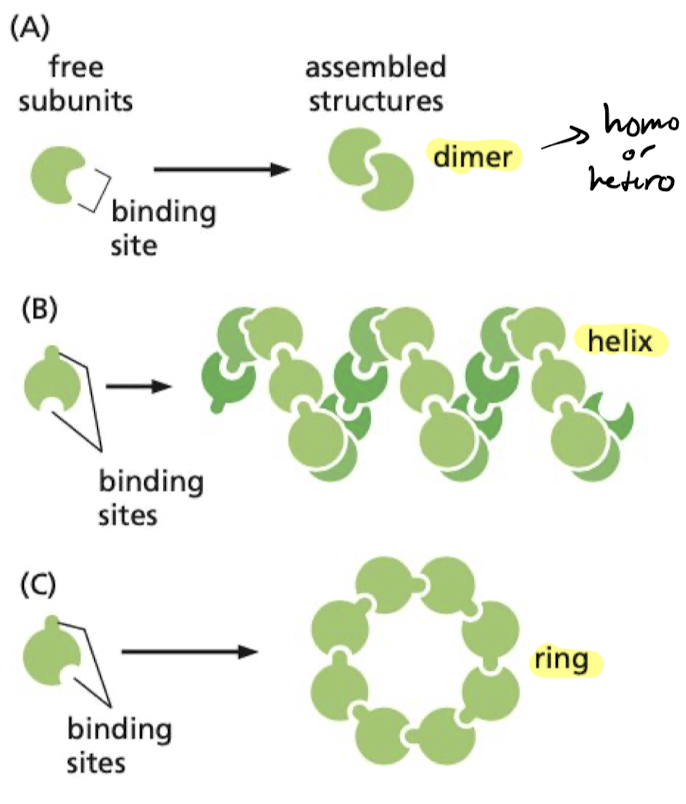
32
New cards
What are the advantages of using smaller protein subunits to build larger structures?
1\. Large structure built from repeating subunits requires minimal genetic information.
2\. Both assembly and disassembly can be readily controlled reversible processes
3\. Errors in the synthesis of the structure can be more easily avoided
* ex) tomato bushy stunt virus (TBSV)
2\. Both assembly and disassembly can be readily controlled reversible processes
3\. Errors in the synthesis of the structure can be more easily avoided
* ex) tomato bushy stunt virus (TBSV)
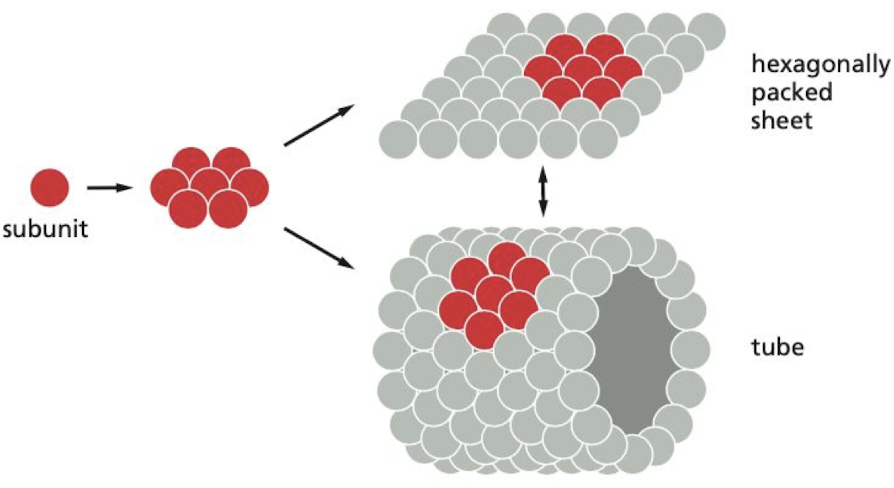
33
New cards
When can protein assemblies go wrong, example?
Amyloid Fibrosis
* beta sheets in brain misfold on top of each other creating giant stacks of proteins
* seen in neurological diseases like parkinson’s and alzheimer’s
* beta sheets in brain misfold on top of each other creating giant stacks of proteins
* seen in neurological diseases like parkinson’s and alzheimer’s
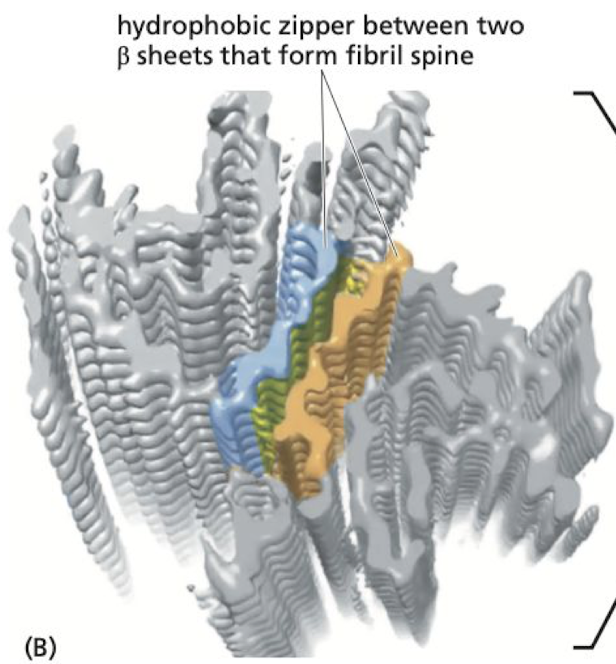
34
New cards
What is a ligand?
The substance that is bound by a protein
35
New cards
How do ligands bind the the binding site of an enzyme?
Weak non-covalent bonds allow proteins selectively bind to a ligand with high affinity.
* Each bond is weak
* Enough bonds form when surface contact between protein and ligand fit very closely.
* “Hand and glove”
* Each bond is weak
* Enough bonds form when surface contact between protein and ligand fit very closely.
* “Hand and glove”
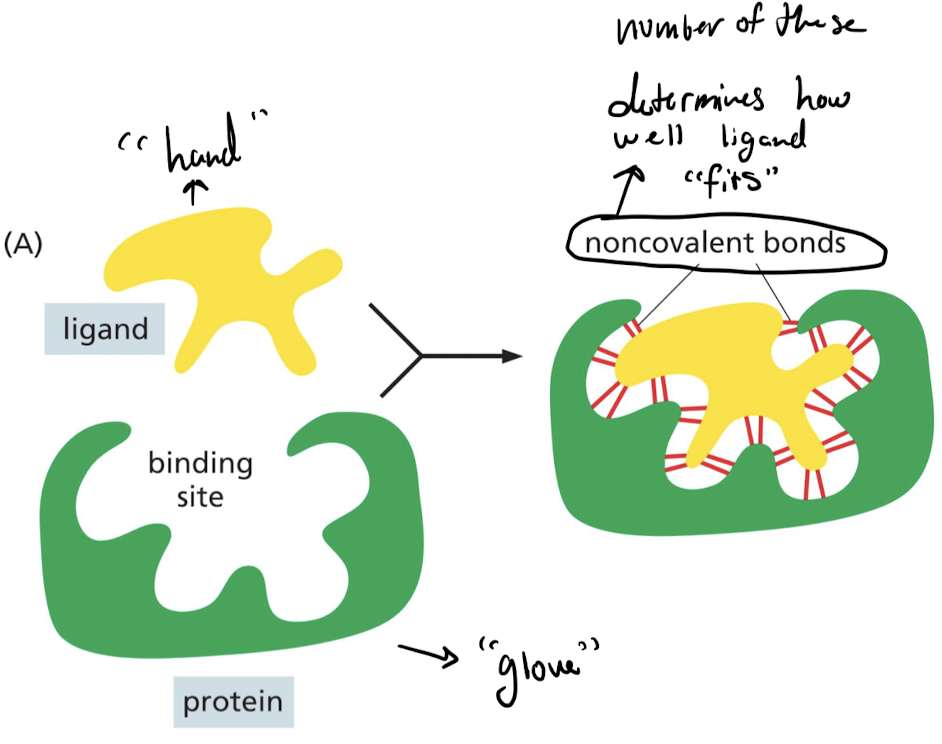
36
New cards
What is a binding site of an enzyme?
The region of a protein that associates with a ligand.
* particular arrangement of amino acids
* amino acids can belong to different portions of the polypeptide chain.
* particular arrangement of amino acids
* amino acids can belong to different portions of the polypeptide chain.
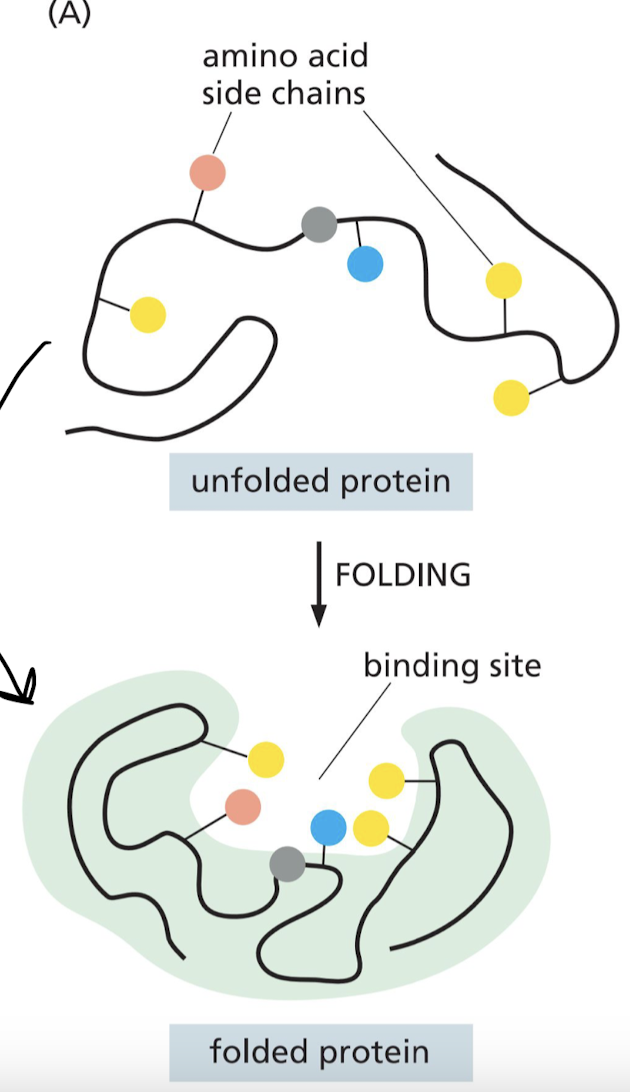
37
New cards
Why are the binding sites of antibodies especially variable?
* Binding site is located between variable domains of heavy and light chains.
* Changes of amino acid length at hypervariable regions leads to enormous diversity
* Changes of amino acid length at hypervariable regions leads to enormous diversity
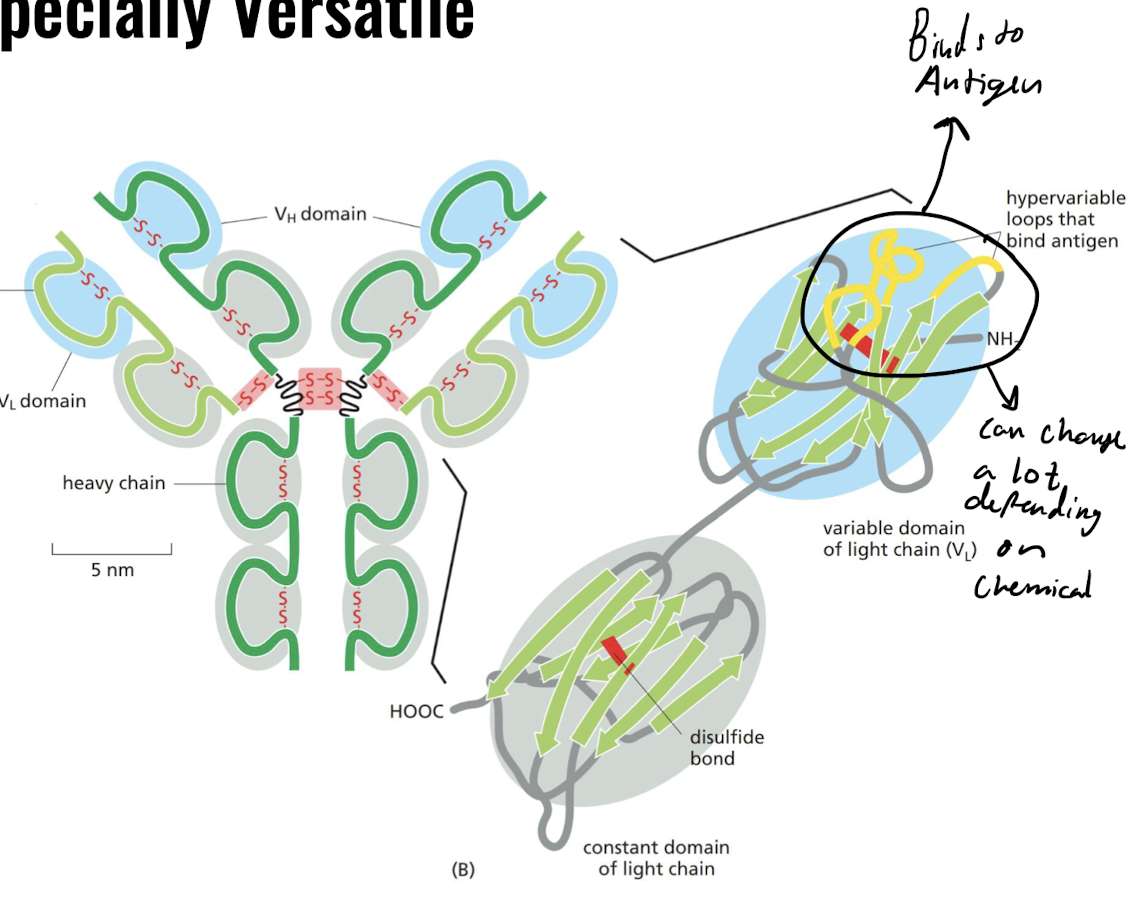
38
New cards
How do enzymes speed up reactions?
* Cause the chemical transformations that make and break covalent bonds.
* selectively stabilizes transition states to lower activation energy
* selectively stabilizes transition states to lower activation energy
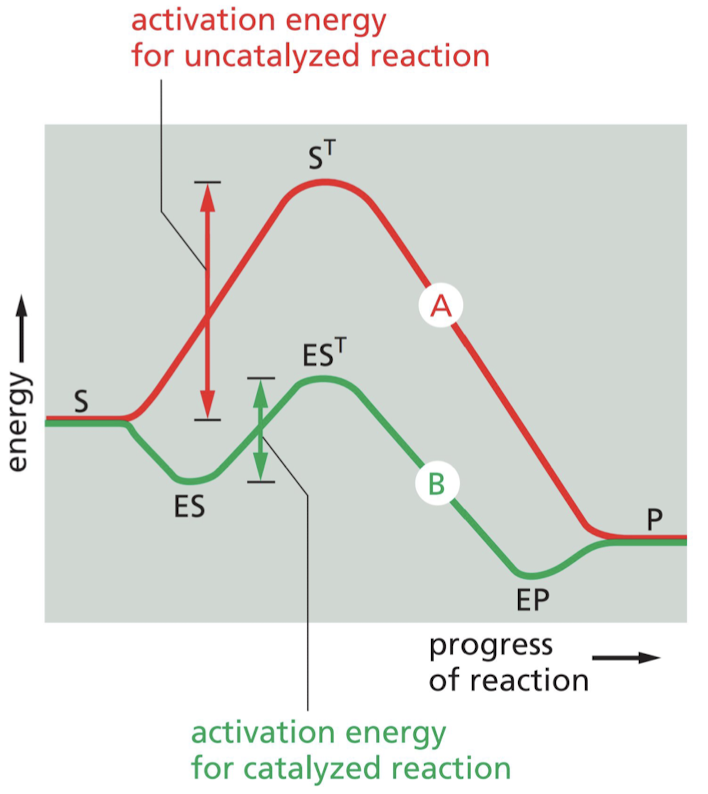
39
New cards
What are the 3 ways that enzymes speed up reactions?
A. binding two substrate molecules together to encourage them to RX with each other
B. rearranging electrons in substrate to create partial negative/positive charges that favor a RX
C. enzyme bends the substrate into a particular shape that forces it into the transition state faster, favoring a RX
B. rearranging electrons in substrate to create partial negative/positive charges that favor a RX
C. enzyme bends the substrate into a particular shape that forces it into the transition state faster, favoring a RX
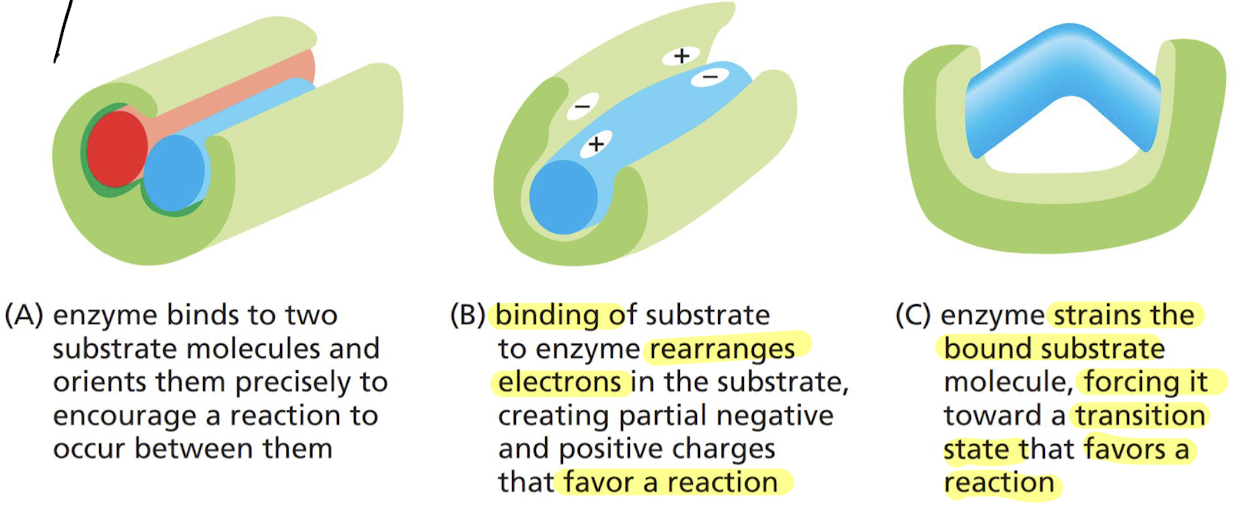
40
New cards
How do enzymes Use Simultaneous Acid and Base Catalysis to speed up reactions?
performs both acid and base catalysis to get fastest RX, with on or the other it is still fast but not as fast as using both.
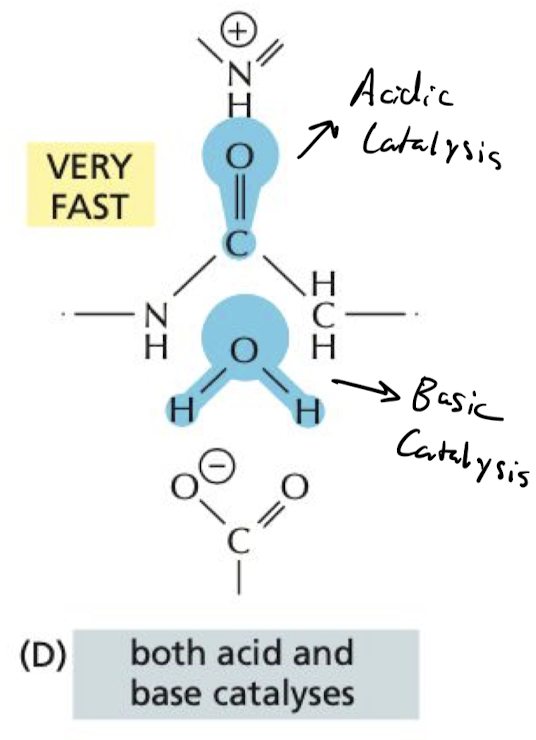
41
New cards
How does the lysozyme enzyme work, and what does it do?
* It catalyzes the cutting of polysaccharide chains in the cell walls of bacteria.
* acts as a natural antibiotic found in saliva, eggs, etc.
* acts as a natural antibiotic found in saliva, eggs, etc.

42
New cards
What are cofactors/coenzymes and whats an example of one?
Cofactors are small non-protein “sidekicks” for enzymes that help them do their function more effectively
* ex) Iron in hemoglobin allows the protein to bind O2 effectively to carry it in RBCs
* ex) Iron in hemoglobin allows the protein to bind O2 effectively to carry it in RBCs
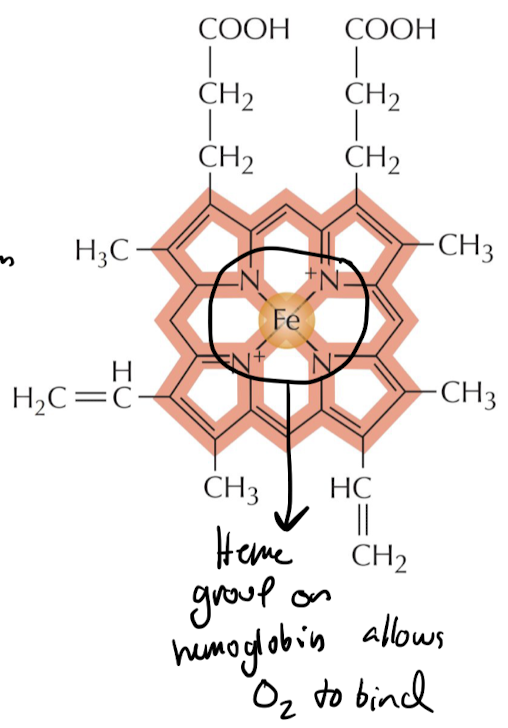
43
New cards
What are different ways that enzyme activity is regulated?
1. enzyme production control
2. isolating enzymes in compartments
3. enzyme degradation control
4. controlling enzyme activity directly (2 ways)
1. allosteric activation/inactivation
2. covalent modification
44
New cards
What is feedback inhibition of enzymes?
Product produced late in a reaction pathway inhibits an enzyme that acts earlier in the pathway.
* example of negative regulation
* example of negative regulation
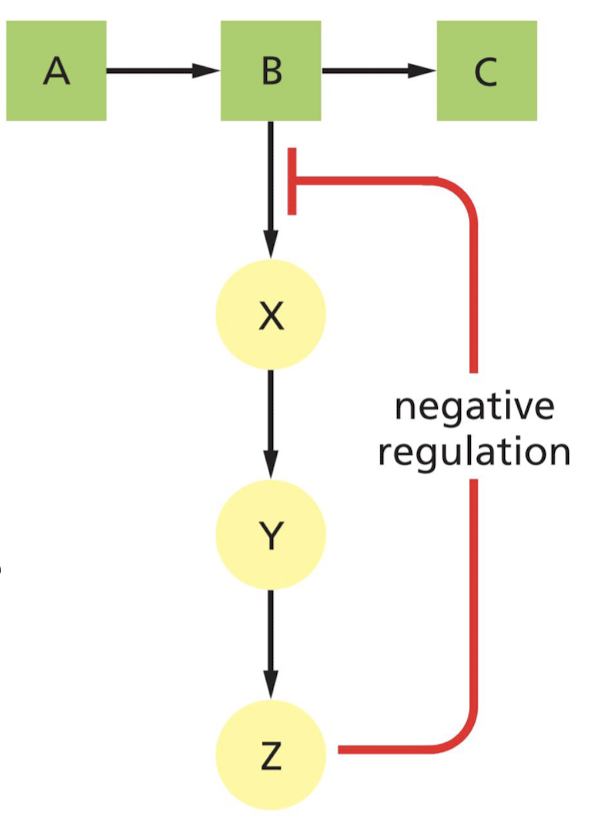
45
New cards
What is the structure of enzymes that implement feedback inhibition regulation?
Enzymes regulated by feedback regulation have two binding sites:
* Active site: binds to substrate
* Regulatory site: binds to regulatory molecule
* Regulatory molecules control the function of the enzyme by conformational change in the protein.
* Active site: binds to substrate
* Regulatory site: binds to regulatory molecule
* Regulatory molecules control the function of the enzyme by conformational change in the protein.
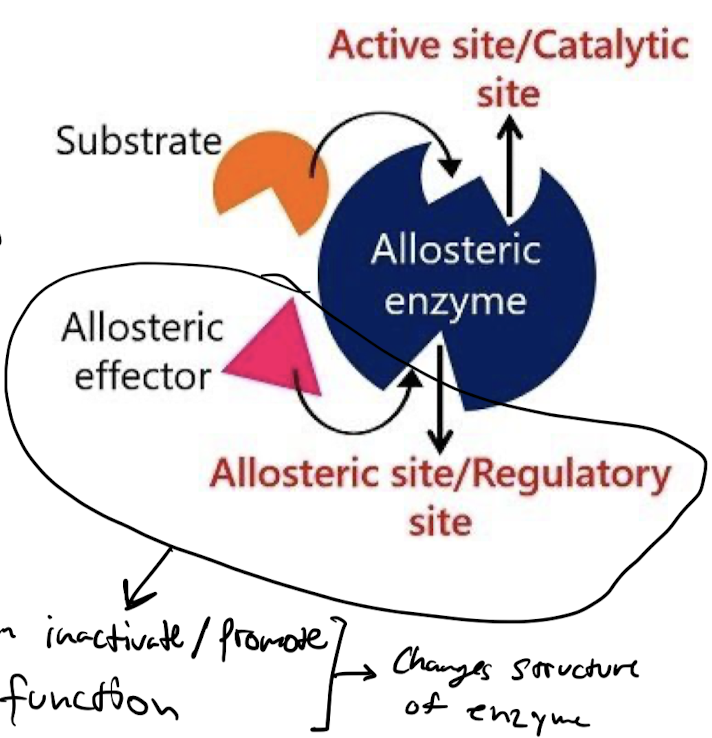
46
New cards
What is positive allosteric regulation of enzymes?
Binding of regulatory molecule **increases** the affinity of the protein to the substrate.
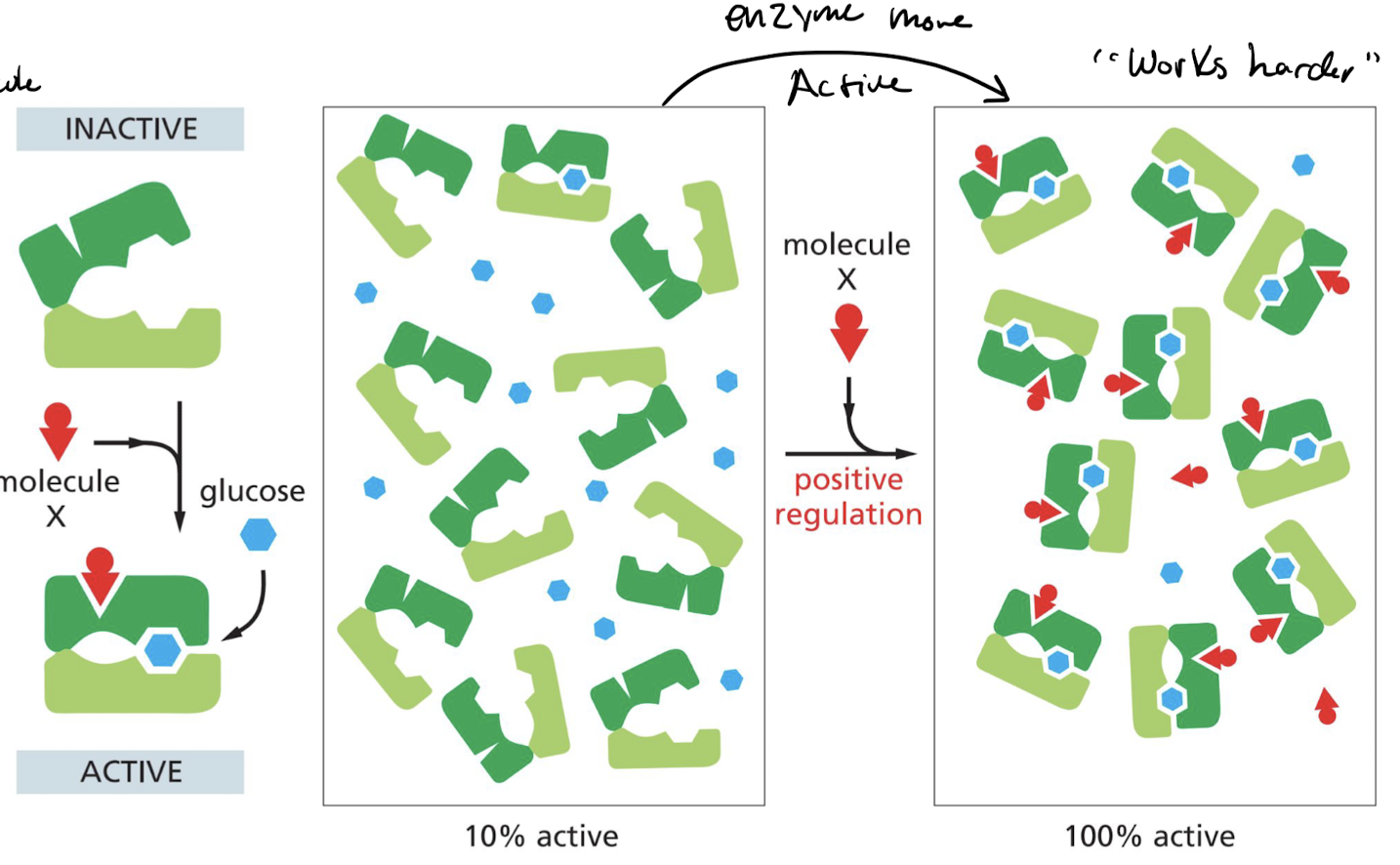
47
New cards
What is negative allosteric regulation of enzymes?
Binding of regulatory molecule **decreases** the affinity of the protein to the substrate.
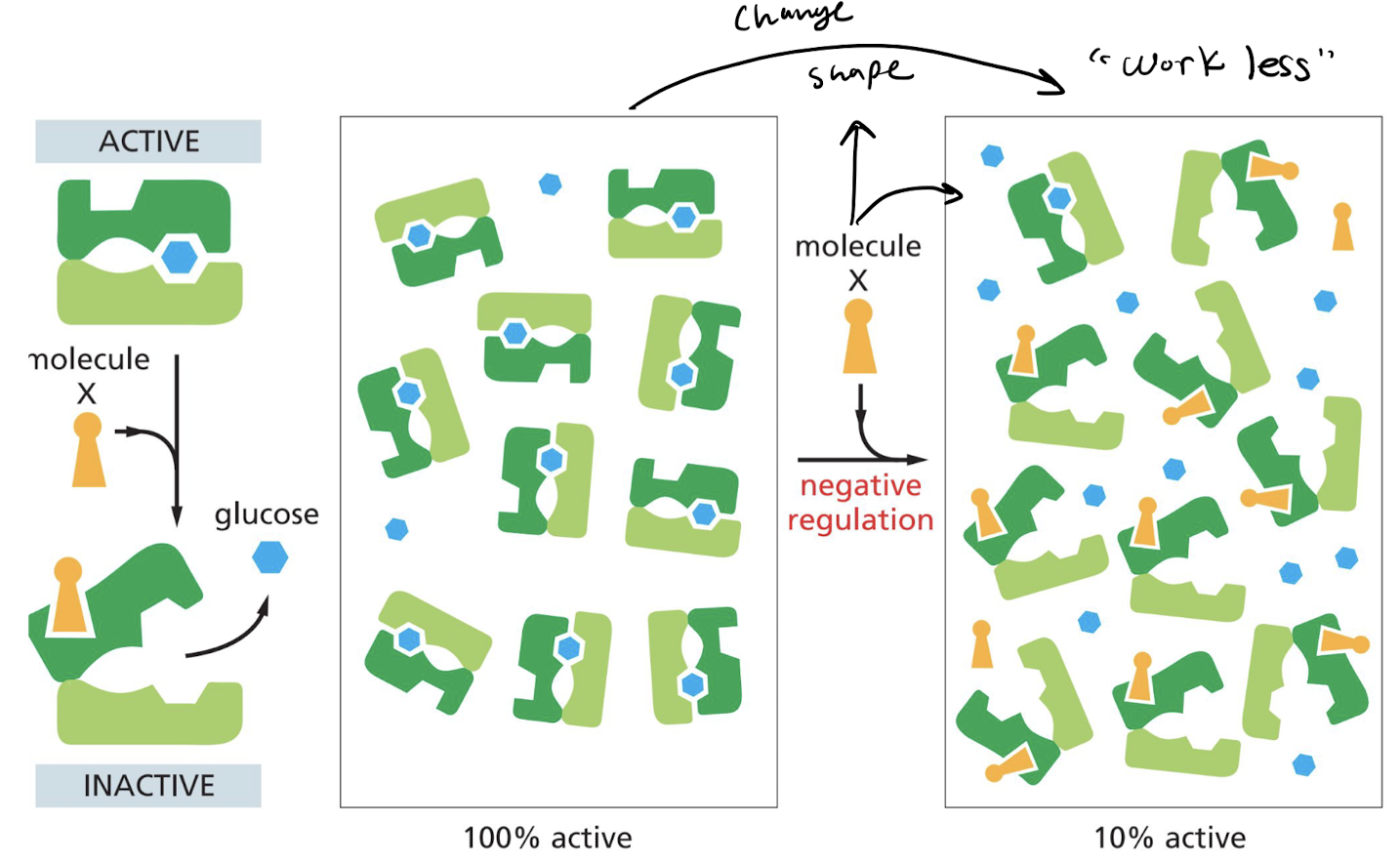
48
New cards
how does protein phosphorylation (via kinase) impact protein function?
* can **bend protein** (conformational change)
* can **create binding sites** for other proteins
* can **disrupt** protein-protein interactions
* effect **can be reversed** by phosphatases
* can either **increase** or **decrease** protein activity
* can **create binding sites** for other proteins
* can **disrupt** protein-protein interactions
* effect **can be reversed** by phosphatases
* can either **increase** or **decrease** protein activity
49
New cards
How is the Src protein regulated by phosphorylation?
* phosphorylated = off
* un-phosphorylated = on
* activating ligand binds to SH3 domain
* phosphorylation in new configuration allows Src to activate
* un-phosphorylated = on
* activating ligand binds to SH3 domain
* phosphorylation in new configuration allows Src to activate
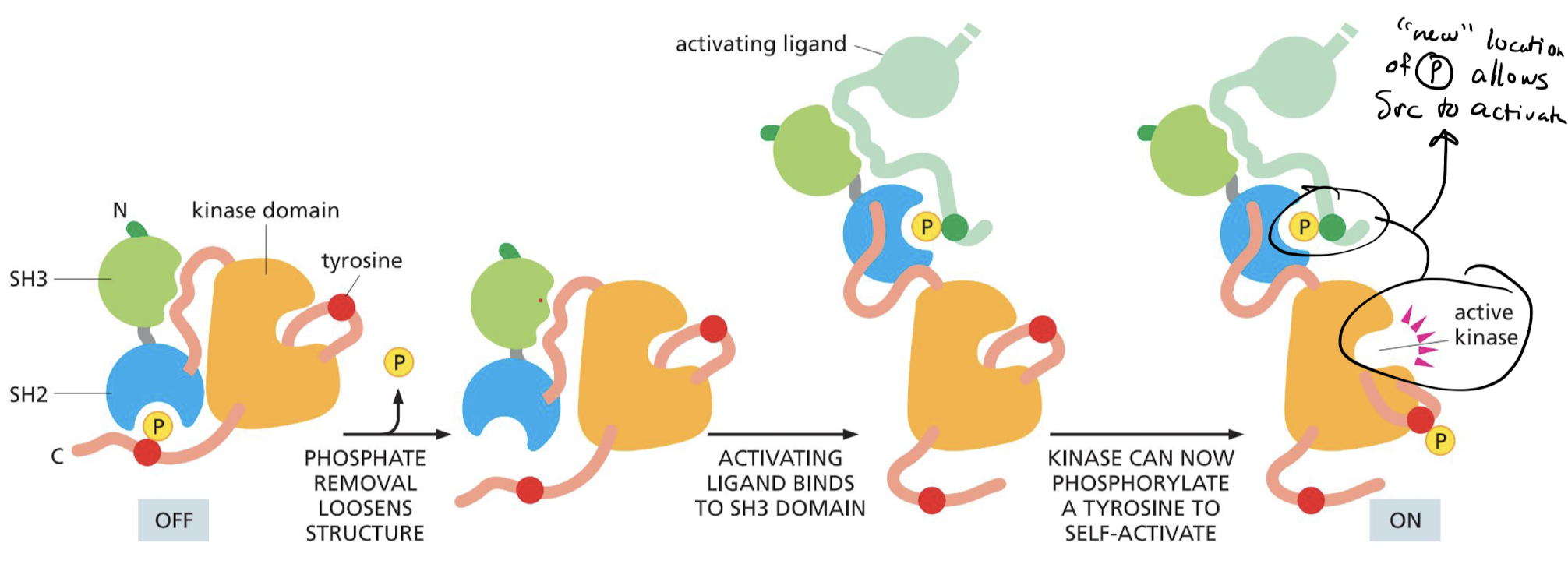
50
New cards
how are GTP binding proteins regulated?
* inactive when phosphate removed = GDP dissociation
* active when phosphate present = GTP binding
* active when phosphate present = GTP binding
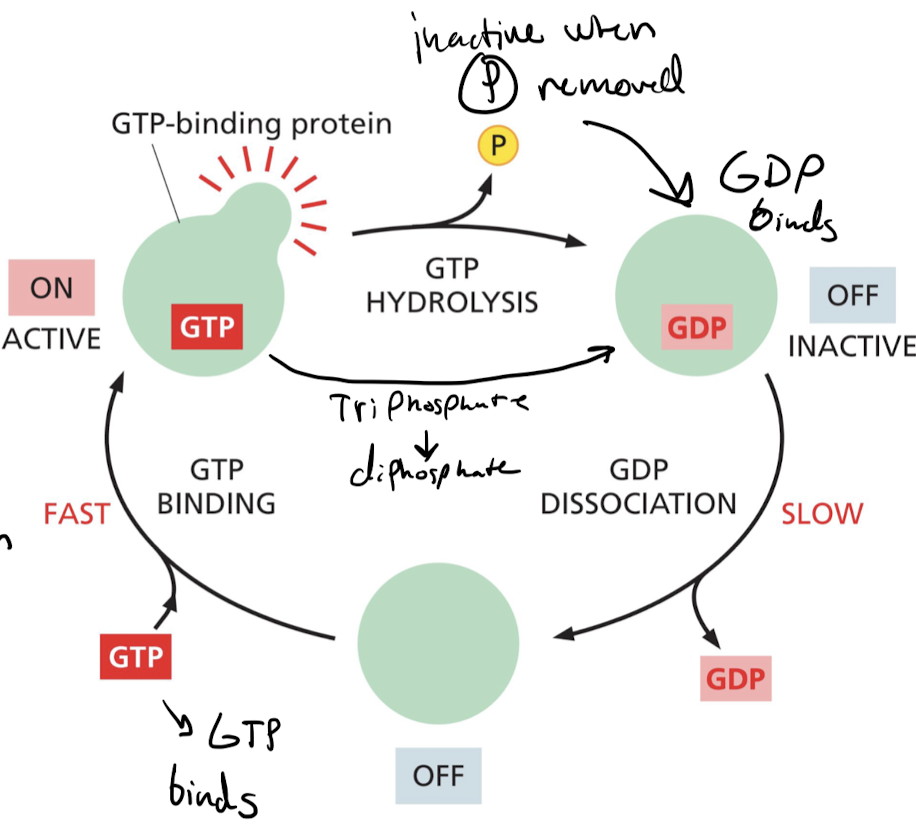
51
New cards
How do motor proteins “walk”?
binding of ATP → ATP hydrolysis → “foot picks up” → release of ADP allows for movement forward
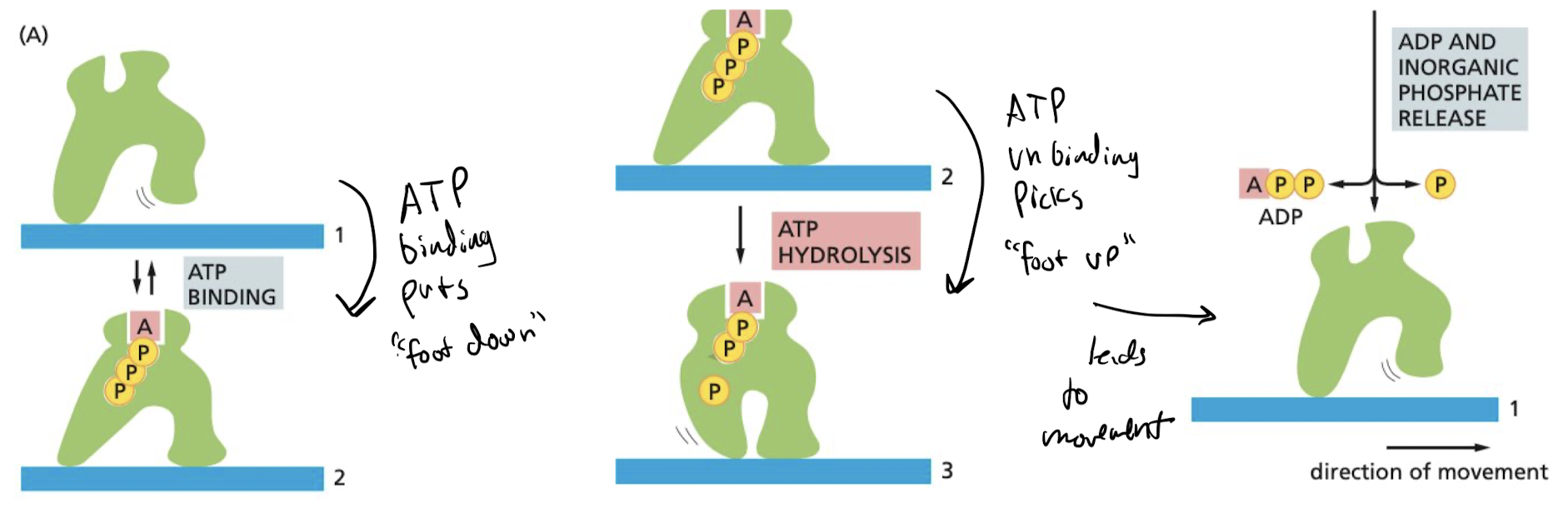
52
New cards
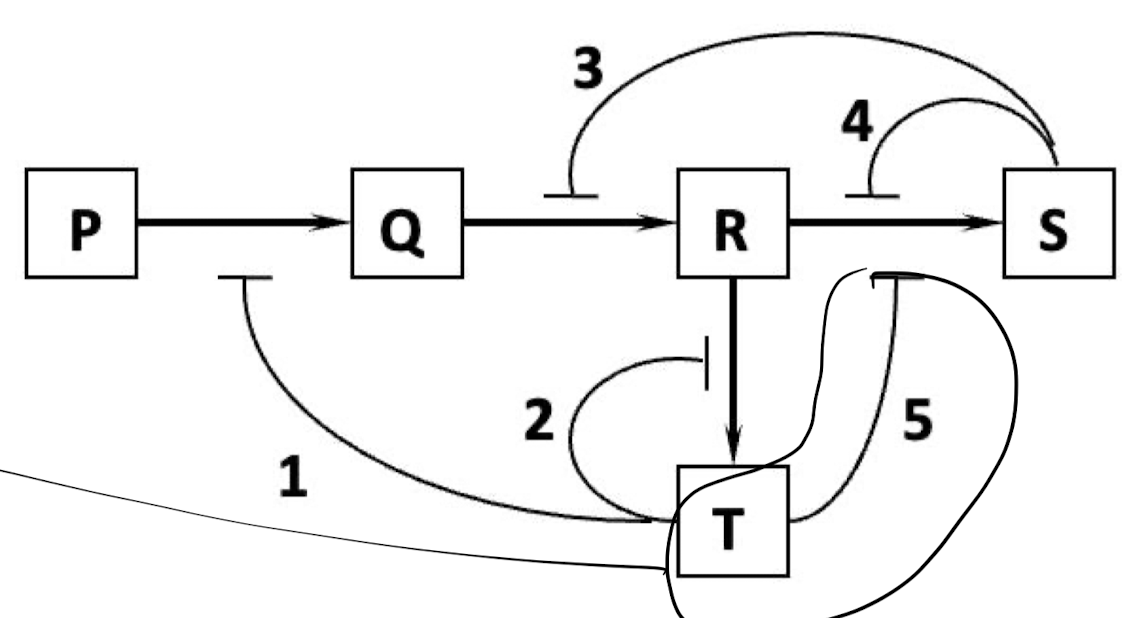
Which of the regulatory interactions 1 to 5 depicted in the following diagram is NOT an example of a negative feedback regulation?
A. 1
B. 2
C. 3
D. 4
E. 5
A. 1
B. 2
C. 3
D. 4
E. 5
E. 5
53
New cards
Two ligands, A and B, bind to two different conformations of the enzyme X. The ligand A is the enzyme’s substrate, whereas ligand B binds to a remote allosteric site and act as the allosteric inhibitor. Which of the following is a consequence of this arrangement?
A. Binding of B to X does not affect the rate of reaction catalyzed by X.
B. Binding of A to X increases the affinity of X for B.
C. Binding of B to X decreases the affinity of X for A.
D. Binding of B to X has a large effect on the binding of A to X, but binding of A to X has a small effect on X–B binding.
A. Binding of B to X does not affect the rate of reaction catalyzed by X.
B. Binding of A to X increases the affinity of X for B.
C. Binding of B to X decreases the affinity of X for A.
D. Binding of B to X has a large effect on the binding of A to X, but binding of A to X has a small effect on X–B binding.
C. Binding of B to X decreases the affinity of X for A.
54
New cards
What was the result of Frederick Griffith’s work on heritable material?
Molecules carrying heritable information are present in the S strain of bacteria.
55
New cards
What were the 4 major molecules that were suspected by Avery, MacLeod, and McCarty to contain heritable information?
1. lipids
2. protein
3. DNA → isolated as source of heritable info
4. RNA
56
New cards
How and who discovered the structure of DNA?
* Double helix initially discovered by **Rosalind Franklin** using X-Ray diffraction analysis
* Structure was later modeled by James Watson and Francis Crick
* Structure was later modeled by James Watson and Francis Crick
57
New cards
What is the basic structure of a DNA molecule
nitrogenous base bound to a sugar, which are bound together by phosphate groups
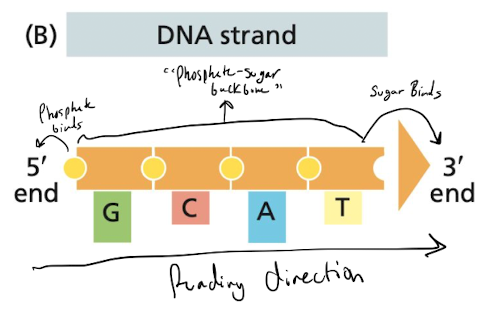
58
New cards
What type of bond links nucleotides together?
phosphodiester bond
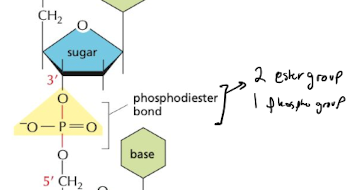
59
New cards
What is the direction of the DNA structure?
antiparallel
60
New cards
Which bases are purine and pyrimidine?
purine = G and A
pyrimidine = C and T
pyrimidine = C and T
61
New cards
how long is the genome stored in a single human cell?
2 meters long
62
New cards
What is a chromosome?
a DNA molecule condensed with proteins
63
New cards
How many chromosomes are in the human genome?
46, 23 from mom and 23 from dad
64
New cards
What is an exon?
segment of eukaryotic genes that codes for proteins
65
New cards
What is an intron?
noncoding region b/w exons
66
New cards
what is a regulatory sequence?
sequence that controls expression of the gene at the proper time, level, and type of cell
67
New cards
What is the basic unit of chromosome structure?
nucleosomes = the first level of compaction
* beads on a string model
* beads = core histones
* string = linker DNA
* beads on a string model
* beads = core histones
* string = linker DNA
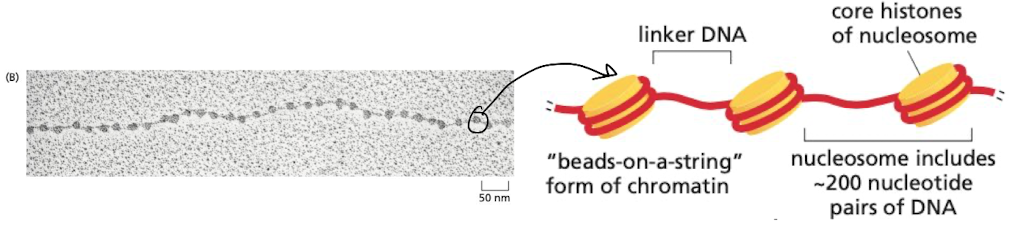
68
New cards
What are the 8 histone proteins (histone octamer) that make up a nucleosome?
1. 2 H2A histones
2. 2 H2B histones
3. 2 H3 histones
1. 2 H4 histones
69
New cards
How does histone H1 help nucleosomal DNA compaction?
* contacts both DNA and the histone octamer and changes path of DNA as it exits nucleosome
* change in exit path helps compaction
* change in exit path helps compaction
70
New cards
What is heterochromatin?
highly condensed form of chromatin, closed and inaccessible
* \~80% of the genome
* the “.zip” fie
* \~80% of the genome
* the “.zip” fie
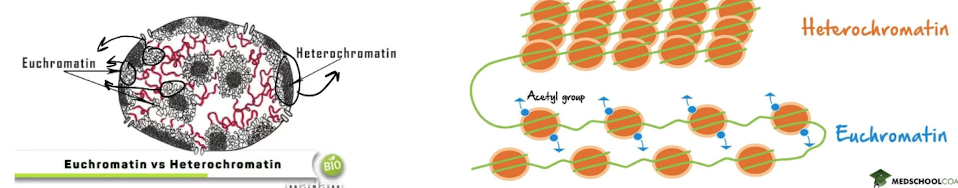
71
New cards
What is euchromatin?
less condensed chromatin, open and readable
* \~20% of genome
* \~20% of genome
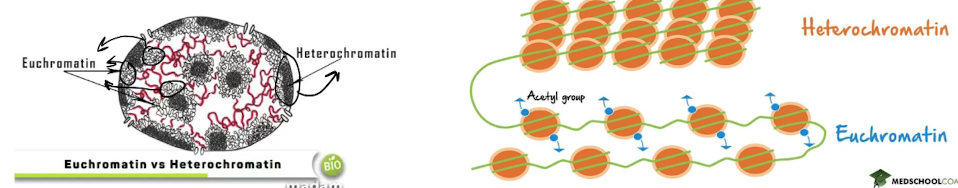
72
New cards
What does nucleosome sliding do?
allows nucleosome to reveal inaccessible parts of DNA
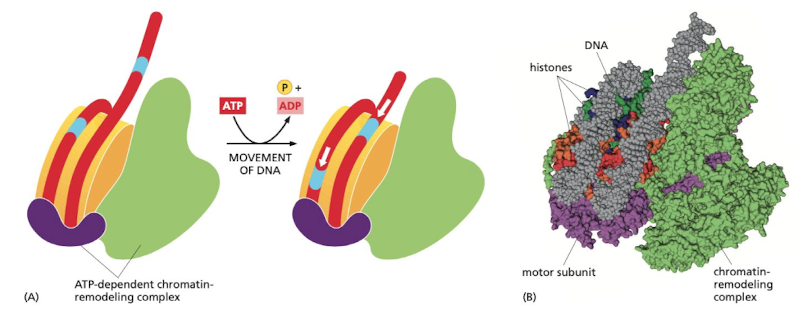
73
New cards
Where do histone modifications occur, are they reversible?
* most modifications occur at N-terminal histone tails
* all types of these modifications are reversible
* all types of these modifications are reversible
74
New cards
What do reader complexes do in the context of histone tails?
they sit on the histone tail to identify regions with changes (damage) and recruits machinery to repair
75
New cards
How do writer enzymes work with reader proteins along histones?
* reader protein recognizes and binds to newly changed nucleosome
* writer enzyme is allosterically activated and attached to the reader
* mechanism works to spread heterochromatin (condensing) message
* writer enzyme is allosterically activated and attached to the reader
* mechanism works to spread heterochromatin (condensing) message
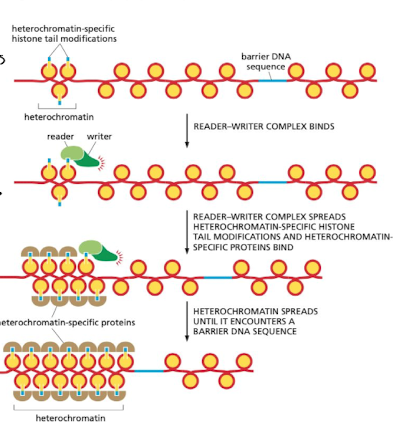
76
New cards
Which feature of DNA underlies its simple replication procedure?
A. The fact that it is composed of only four different types of bases
B. The antiparallel arrangement of the double helix
C. The complementary relationship in the double helix
D. The fact that there is a major groove and a minor groove in the double helix
A. The fact that it is composed of only four different types of bases
B. The antiparallel arrangement of the double helix
C. The complementary relationship in the double helix
D. The fact that there is a major groove and a minor groove in the double helix
C. The complementary relationship in the double helix
77
New cards
Each chromosome in a human eukaryotic cell is composed of how many independent double strands of DNA?
A. 1
B. 2
C. 23
D. 46
E. Over 200
A. 1
B. 2
C. 23
D. 46
E. Over 200
A. 1
78
New cards
What is transcription?
Process of copying DNA information into a RNA molecule.W
79
New cards
What is translation?
Process where mRNA is used to direct the synthesis of a protein.
80
New cards
What are the differences between DNA and RNA?
1. nucleotides in RNA are ribonucleotides
2. RNA has U instead of T
3. RNA is usually single-stranded
4. RNA molecules can fold into 3D shapes
1. allows for enzymatic function (ribosomes)
81
New cards
What are the 3 steps of transcription?
1. DNA is unwound and bases are exposed
2. one strand serves as template for RNA synthesis
3. RNA is synthesized
82
New cards
Which DNA strand is the template strand and which is the coding strand?
5’ - 3’ = coding strand
3’ - 5’ = template strand → used in transcription
3’ - 5’ = template strand → used in transcription
83
New cards
What is the process of transcription like in prokaryotes?
1. RNA pol holoenzyme finds promoter (sigma factor)
2. RNA polymerase holoenzyme forms transcription bubble at promoter
3. abortive initiation
4. promoter is cleared and sigma factor is released
5. elongation
6. termination → formation of hairpin at terminal signal (AT rich region)
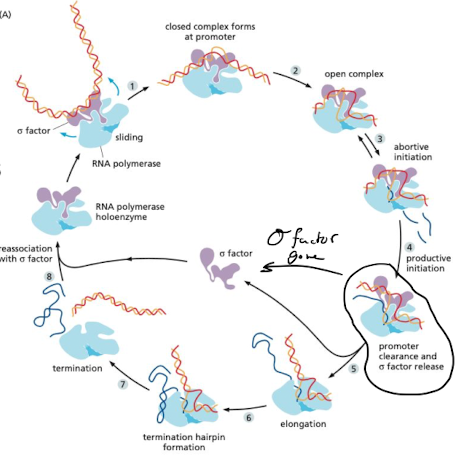
84
New cards
What are required in order for eukaryotes to initiate transcription?
general transcription factors = allow RNA pols to find promoters and begin transcription
85
New cards
What happens in initiation of transcription in eukaryotes?
TBP subunit of TFIID binds to TATA box 30 nucleotides before the promoter
* causing a distortion of DNA and acts as a marker for active promoter
* more general TFs assemble alongside RNA Pol II to form **transcription initiation complex**
* TFIIH separates two DNA strands, exposing template
* phosphorylates CTD of RNA pol II allowing it to be released from general TFs → begins elongation
* causing a distortion of DNA and acts as a marker for active promoter
* more general TFs assemble alongside RNA Pol II to form **transcription initiation complex**
* TFIIH separates two DNA strands, exposing template
* phosphorylates CTD of RNA pol II allowing it to be released from general TFs → begins elongation
86
New cards
What proteins are “also” needed for initiation of transcription?
* **Activator Protein** = binds to enhancer and interacts with transcription initiation complex via mediator protein complex
* **Chromatin-modifying enzymes**
* **Chromatin-modifying enzymes**
87
New cards
Where is polycistronic mRNA found?
**Only** in bacteria
88
New cards
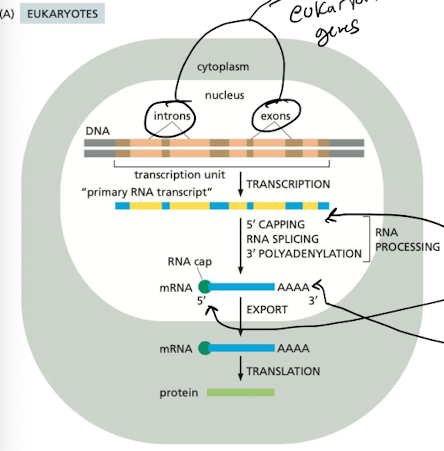
What are the steps of RNA processing in eukaryotes?
1. 5’ capping
2. intron splicing
3. 3’ polyadenylation
89
New cards
How does 5’ capping occur?
* phosphorylation of RNA Pol II CTD recruits proteins initiating RNA processing
* modified guanine nucleotide (7-methylguanosine) attached to 5’ end of RNA, contains new 5’ to 5’ linkage
* cap acts as a signal for packaging, protecting the 5’ end of the RNA
* modified guanine nucleotide (7-methylguanosine) attached to 5’ end of RNA, contains new 5’ to 5’ linkage
* cap acts as a signal for packaging, protecting the 5’ end of the RNA
90
New cards
How are introns spliced during eukaryotic RNA processing?
* **spliceosome** carries out transesterification
* 5’ end snipped first, followed by 3’ end
* cut out intron = **lariat**
* 5’ end snipped first, followed by 3’ end
* cut out intron = **lariat**
91
New cards
How is 3’ polyadenylation carried out during eukaryotic RNA processing?
* CstF and CPSF bind to phosphorylated CTD on RNA Pol II
* these 2 factors recognize and bind to signal sequence on 3’ end of mRNA
* CsF and CPSF recruit additional factors that cut downstream of sequence
* PAP binds to freshly cleaved end and adds \~200 As to the end
* these 2 factors recognize and bind to signal sequence on 3’ end of mRNA
* CsF and CPSF recruit additional factors that cut downstream of sequence
* PAP binds to freshly cleaved end and adds \~200 As to the end
92
New cards
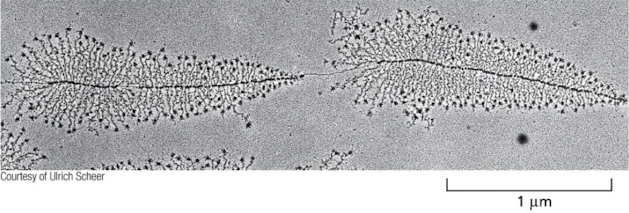
Due to their high transcription rate, active ribosomal RNA (rRNA) genes can be easily distinguished in electron micrographs of chromatin spreads. They have a characteristic “Christmas tree” appearance, where the DNA template is the “trunk” of the tree and the nascent RNA transcripts form closely packed “branches.” At the base of each branch is an RNA polymerase extending that branch, while RNA processing complexes at the tip of the branch form terminal “ornaments.” Would you expect the top of the tree to represent the beginning or end of the rRNA gene? Would the “ornaments” be at the 3′, 5′, or both ends of the nascent rRNA molecules?
a. End; 3′
b. End; 5′
c. Beginning; either 3′ or 5′
d. Beginning; 3′
e. Beginning; 5′
a. End; 3′
b. End; 5′
c. Beginning; either 3′ or 5′
d. Beginning; 3′
e. Beginning; 5′
e. Beginning; 5’
93
New cards
This large and complex general transcription factor has a DNA helicase activity that exposes the template for RNA polymerase II transcription. It also has a kinase activity that phosphorylates the C-terminal domain of the polymerase on Ser5 leading to promoter clearance. What is it?
a. TFIIB
b. TFIID
c. TFIIE
d. TFIIF
e. TFIIH
a. TFIIB
b. TFIID
c. TFIIE
d. TFIIF
e. TFIIH
e. TFIIH
94
New cards
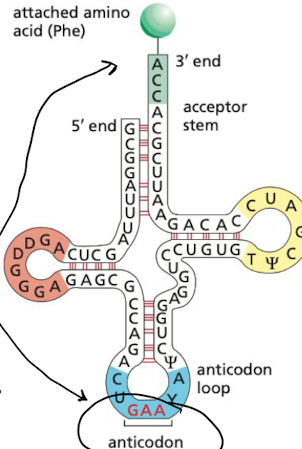
what are transfer RNAs (tRNAs)?
adaptor molecules that can recognize and bind both to the codon and to the amino acid
95
New cards
What are anticodons?
set of thee consecutive nucleotides that pair with complementary codon in an mRNA molecule
96
New cards
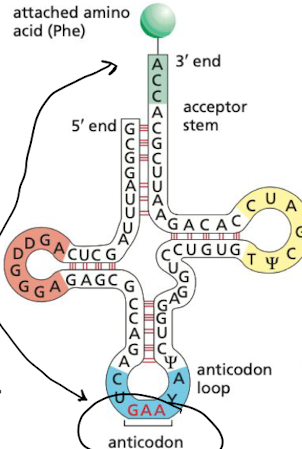
What is the amino acid attachment site?
short single strand region on 3’ end allows for codon to attach
97
New cards
What is the “wobble” position between a codon and anticodon?
it is where the 3rd nucleotide would match up, weaker bond. Because first 2 nucleotides are usually same for most amino acids
98
New cards
What do aminoacyl-tRNA synthetases do?
covalently couple each amino acid to its appropriate set of tRNA molecules → makes it **charged** and primed for translation
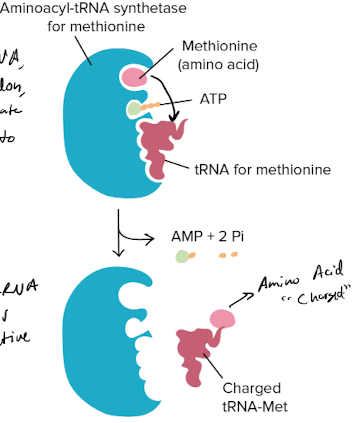
99
New cards
Where are new amino acids added along a growing chain of amino acids of a protein?
along the C-terminal end
* goes from N → C
* goes from N → C
100
New cards
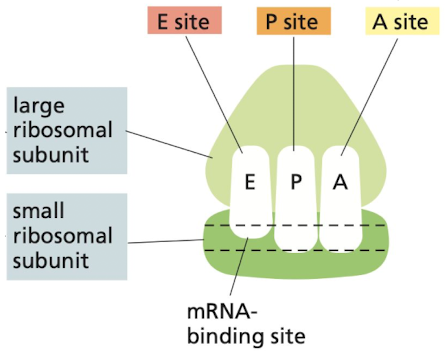
What are the 3 sites that tRNAs bind to within the ribosome?
1. A site: binds to charged tRNA
2. P site: holds tRNA with growing polypeptide chain
3. E site: exit site