RRoy2: Eukaryotic Transcription
1/16
Earn XP
Description and Tags
2nd rick roy lecture
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
17 Terms
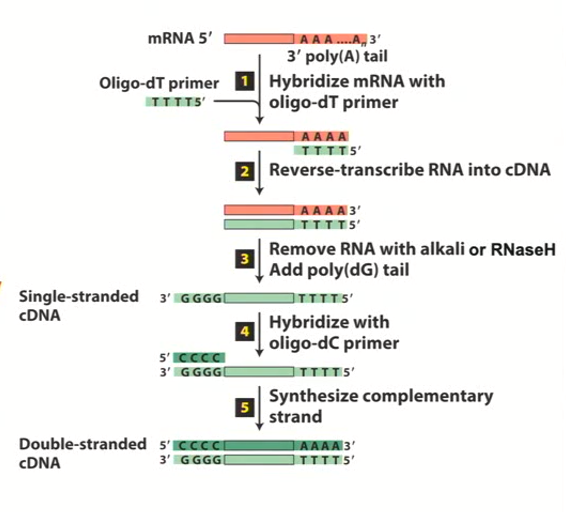
cDNA library formation using Reverse Transcriptase (RT)
complementary DNA is synthesized from mRNA by priming the poly A tail with a single-stranded Poly-T oligonucleotide.
Reverse Transcriptase uses this primer to initiate single strand DNA synthesis that is complimentary to mRNA template
RNA is then removed and a poly dG adapter is annealed to the 3’ end
A poly dC primer is used to initiate synthesis of the second DNA strand
E. coli DNA polymerase I progresses through any remaining hybrid regions and extends to the second strand
The result is a double-stranded cDNA molecule that represents the original mRNA. This cDNA can be cloned into vectors for further analysis or expression.
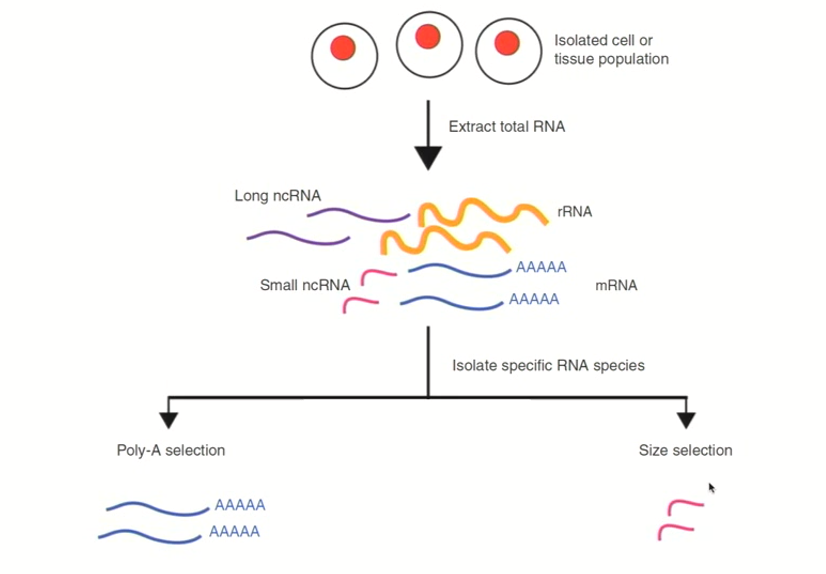
RNA-seq starting process
you get an RNA sample from any tissue/source you might be interested in
This will give a complex mix of RNAs which you can separate out using affinity chromatography (run all this RNA over a poly-DT column, & collect all the mRNA because the poly-A tails will interact with the poly-DT oligonucleotides)
All of the mRNA will then be converted into double stranded cDNAs
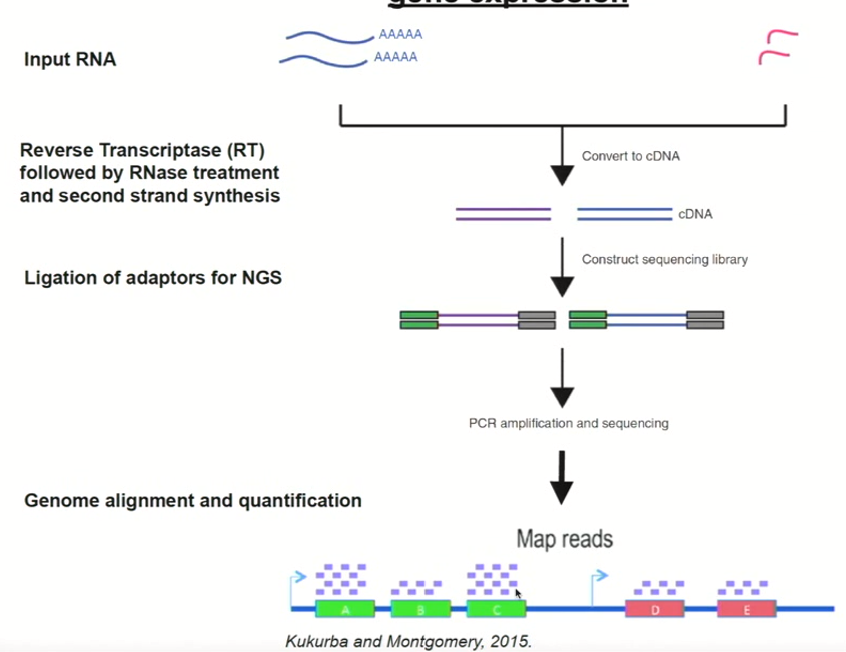
RNA-seq cDNA adapting
After mRNA is converted to double stranded cDNA, we can adapt the cDNAs
We do this by putting on known sequence adaptors at each end of the cDNA molecules throughout the entire sample
Then, we can amplify using PCR and sequence with next generation sequencing
We have sequence information that is representative of how often a given cDNA was present in the sample
you can take this information and realign it to the genome sequence we already have (reference)
We have an idea of how many reads corresponded to every single gene in the genome (aka. which genes were expressed and how strongly)
RNA-seq is the go to technique to understand how genes are being expressed in organisms
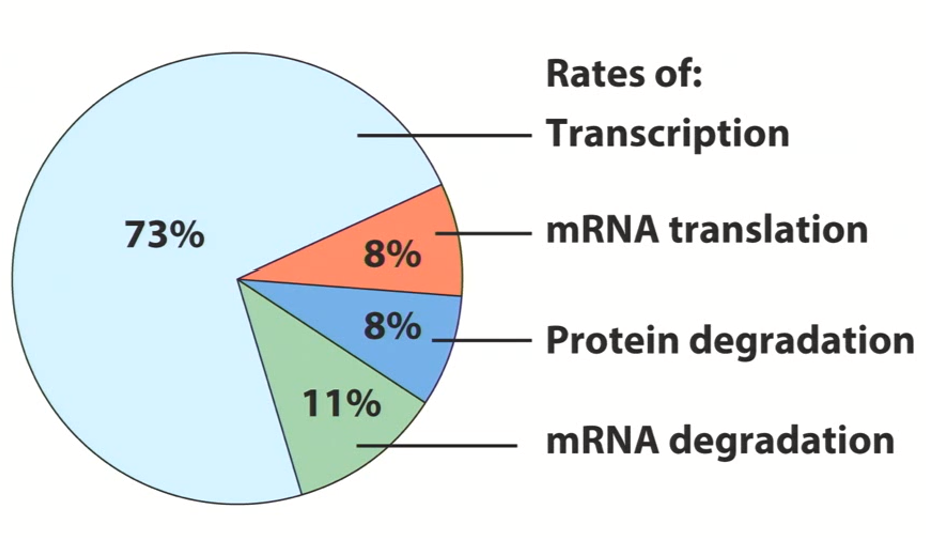
What is the major regulator of gene expression?
Transcriptional Regulation
Overview of Transcription
DNA double helix locally denatures, one strand acts as a template
rNTPs (nucleotides) are incorporated complementary to the template strand
When the rNTPs come into the catalytic region of the RNA polymerase, it takes the nucleotide and incorporates the alpha phosphate into the growing polymer chain by releasing it from the beta phosphate

Conventions for describing RNA Transcription (including start site, downstream, upstream, and non template strand)
Start site is where RNA polymerase makes its first phosphodiester bond (it is always considered +1)
Coding sequence is downstream from the start site and will usually include a translational start site (AUG) in the mRNA
Elements that regulate the initiation of transcription are upstream (known as the promoter - has all kinds of information that tells RNA polymerase to attach at the start site)
Non-template strand is the second DNA strand that is not read by RNA polymerase, but has an identical sequence (except Ts are Us instead)
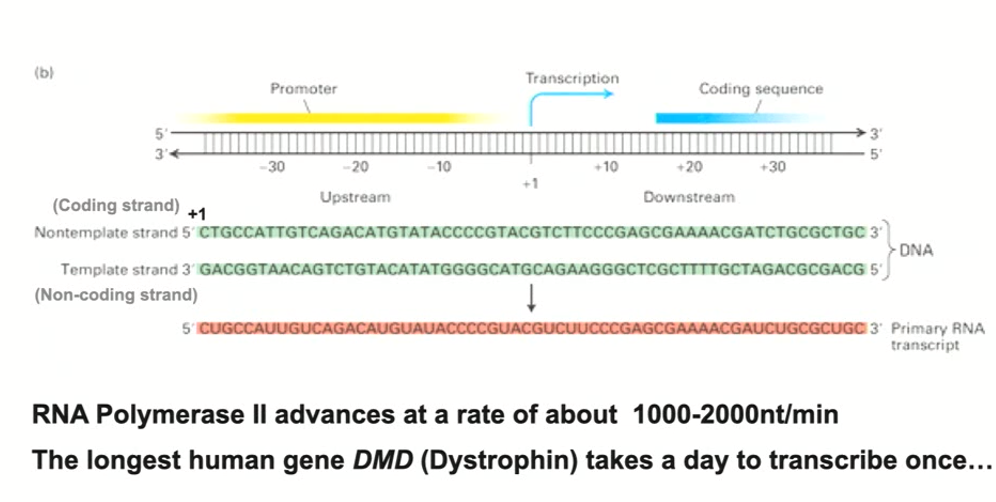
How fast does RNA polymerase transcribe?
1000-2000 nucleotides/minute
What are the 3 stages of transcription?
Initiation
RNA polymerase locally unwinds, and RNA polymerase binds to the promoter to start synthesizing
Elongation
Polymerase moves along the DNA template in the 3’→5’ direction, adding nucleotides in the synthesized strand in the 5’ → 3’ direction.
Termination
RNA polymerase receives a stop signal and is released from the RNA
Similarities between prokaryotic and eukaryotic transcription (lactose)
When lactose is present, it allosterically binds to the lac repressor, detaching it from the DNA
This allows the polymerase to now bind to the DNA and transcribe
RNA polymerase uses this mechanism to regulate the rate of transcription
In summary, in high lactose environments, RNA polymerase transcribes much faster
Other auxillary factors, such as cAMP+CAP, also regulate rate of transcription

Characteristics of prokaryotic transcription not found in eukaryotic transcription
In prokaryotic transcription, 1 single mRNA may contain several different units that will be independently translated (these gene products are organized into operons)
Polymerase makes one big mRNA that has different gene products separated by linker sequences that have their own ribosomal recognition sites (Polycistronic)
As the mRNA is being made, ribosomes interact with these sites and make the proteins as it’s being transcribed.
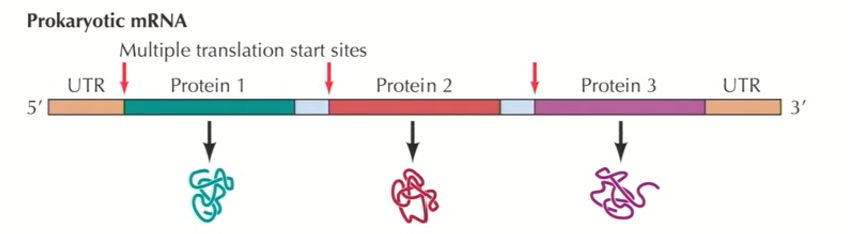
Characteristics of eukaryotic transcription not found in prokaryotic transcription
much more complex mRNA structures (have a cap)
untranslated regions upstream of the start site
Another untranslated region (after the sequence that codes for the protein)
A poly-A tail
Usually, one transcription unit gives rise to one protein (Monocistronic)
3 RNA Polymerases found in eukaryotic transcription
RNA polymerase 1
RNA polymerase 2
RNA polymerase 3
RNA polymerase 1
Its distinct substrates and products that it makes are associated with ribosomal components
It is present in the nucleolus
It is critical for making ribosomal RNA. It synthesizes the precursor of ribosomal RNA (rRNA) and is essential for ribosome biogenesis.
RNA polymerase 3
critical for transfer RNA (tRNA)
it also makes one RNA component known as the 5S rRNA
It also makes one splicing RNA called snRNA U6
RNA polymerase 2
the enzyme that makes mRNAs (protein-coding genes)
Its responsible for the formation of messenger RNA (mRNA), small nuclear RNAs (snRNAs), Small interfering RNAs (siRNAs) and Micro RNAs (miRNAs)
RNA polymerases all look like their bacterial ancestor (evolutionary conserved) structures look very similar
Colours in the image are meant to represent structures that share some sort of conformation
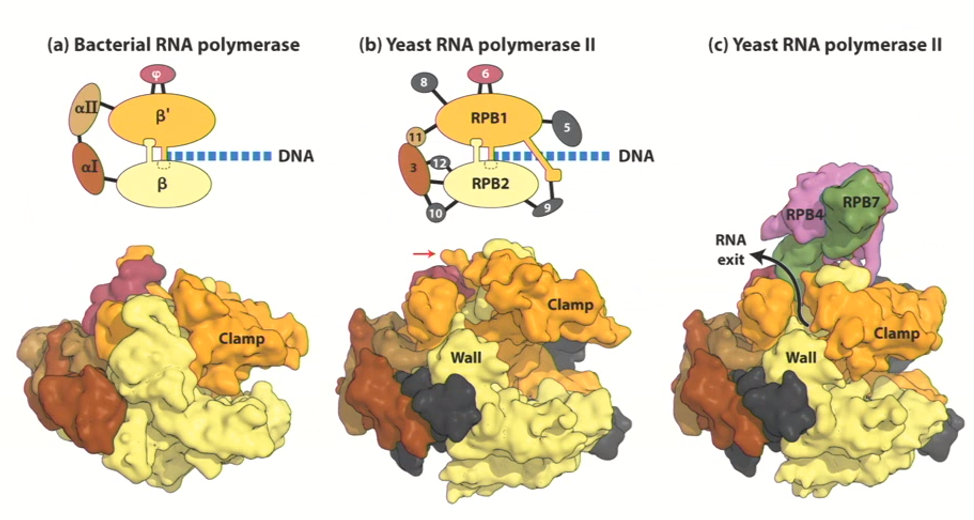
Eukaryotic RNA Polymerase II special structure
It has a clamp that changes conformation during elongation
Once RNA polymerase interacts with DNA and starts to form a product, the clamp comes down over the DNA
This prevents the RNA from detaching from DNA (and why RNA polymerase transcribes so fast)
CTD on RNA polymerase II
Carboxy Terminal Domain of RNA polymerase II (unique to this polyemerase)
Heptapeptide sequence that gets repeated over and over again
In humans it is repeated 52 times
It is completely unstructured, which is odd