Biochem - 5.1/5.2 (lect + book notes)
1/189
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
190 Terms
Q1: Who developed the first method to deduce amino acid sequence of short polypeptides, and what reagent was used?
A1: Frederick Sanger developed the method using 1-fluoro-2,4-dinitrobenzene.
Q2: What was the principle behind Sanger’s protein sequencing method?
A2: Chemical modification of the amino-terminal residue with 1-fluoro-2,4-dinitrobenzene, followed by acid hydrolysis and identification via chromatography.
Q3: What was a limitation of Sanger's method?
A3: Acid hydrolysis destroyed the sample, requiring new protein for each reaction.
Q4: Who improved Sanger’s method and how?
A4: Pehr Edman replaced 1-fluoro-2,4-dinitrobenzene with phenylisothiocyanate (PITC) for N-terminal tagging.
Q5: How does Edman degradation work (steps)?
A5: PITC reacts with the N-terminus; trifluoroacetic acid cleaves the first amino acid; it's extracted, converted to a PTH derivative, and identified via chromatography.
Q6: What's a major advantage of Edman degradation over Sanger’s method?
A6: No need for additional protein in each cycle.
Q7: What's a limitation of Edman degradation?
A7: Inefficiencies accumulate, limiting it to relatively short peptide sequences.
Q8: How many amino acids can Edman degradation sequence using <1 µg sample?
A8: Up to 50 amino acid residues.
Q9: Why must proteins be cleaved before Edman degradation?
A9: It can’t sequence polypeptides longer than ~50 residues in one run.
Q10: How are proteins typically cleaved for sequencing?
A10: Using proteases like trypsin (cleaves after K/R) and chymotrypsin (cleaves after Y, W, F, and less efficiently L, M).
Q11: What is a strategy for reconstructing the full sequence after cleavage?
A11: Generating overlapping fragments and reassembling based on shared sequences.
Q12: What are some alternative cleavage agents to trypsin/chymotrypsin?
A12: Staphylococcus aureus V-8 protease (cleaves after D, E) and cyanogen bromide (cleaves after M).
Q13: What does mass spectrometry measure in peptides?
A13: Mass-to-charge ratio (m/z), used to determine molecular mass.
Q14: What fundamental physics principle underlies mass spectrometry?
A14: Newton’s 2nd law: F=maF = maF=ma; applied force = mass × acceleration.
Q15: What are the two ionization methods used for peptides?
A15: Electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI).
Q16: Describe electrospray ionization (ESI).
A16: Releases charged peptide fragments via high voltage, causing solvent to evaporate.
Q17: Describe MALDI.
A17: Uses a laser to ionize a matrix, transferring charge to embedded proteins or fragments.
Q18: How is ion acceleration measured in a mass spectrometer?
A18: Acceleration is measured along a curved path to calculate mass.
Q19: What technique determines protein identity using two mass spectrometers?
A19: Tandem mass spectrometry (MS/MS).
Q20: What happens in tandem MS?
A20: MS1 selects peptide fragments, a collision chamber breaks them, MS2 measures subfragment masses.
Q21: How is the amino acid sequence deduced via tandem MS?
A21: By comparing experimental peptide masses to predicted tryptic fragments in genome databases.
Q22: Why is trypsin commonly used before MS analysis?
A22: It cleaves after K/R, allowing mass comparisons to predicted tryptic fragments.
Q23: How are high-probability matches from MS verified?
A23: Using protein biochemistry methods.
Q24: What is SPPS used for?
A24: Synthesizing peptide antigens and peptide-based therapeutic drugs.
Q25: How is SPPS directionally opposite to in vivo synthesis?
A25: SPPS adds amino acids from C to N terminus; in vivo adds from N to C terminus.
Q27: What’s the basic mechanism of SPPS (Dev by Bruce Merrifield)?
A27: One amino acid at a time is added via covalent linkage to the growing peptide chain.
Q28: What are the five steps of SPPS?
A28:
Step 1: Attach C-terminal AA to resin; remove Fmoc.
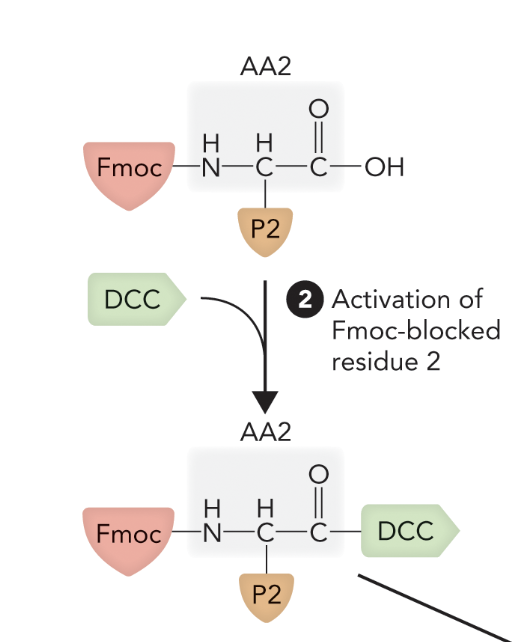
Step 2: Activate next AA with DCC, add to column.
Step 3: Efficient coupling between AAs; remove unreacted compounds.
Step 4: Remove protecting groups.
Step 5: Release peptide from resin via hydrogen fluoride.
Q29: What prevents unwanted side reactions in SPPS?
A29: Protecting groups on amino acid side chains.
Q30: What is the efficiency and scale of SPPS?
A30: Automated; can produce 15–25 amino acid peptides in hours using <1 µg material.
What are the pros and cons of identifying an unknown protein using Edman degradation vs tandem mass spectrometry?
Edman Degradation
✅ Advantage: Directly sequences N-terminal amino acids using PITC labeling and chromatography.
❌ Disadvantage: Only sequences from the amino-terminal end; covers a small region of the protein.
Mass Spectrometry
✅ Advantage: Analyzes complex mixtures without purification; faster sample prep; uses genome databases for ID.
❌ Disadvantages: Requires genome database access; identification is probabilistic, not direct.
What happens if you forget to protect amino acid 2 during solid-phase peptide synthesis of DALTAEMS?
Without the Fmoc group on amino acid 2, multiple side reactions can occur. It may couple incorrectly, leading to unwanted or branched peptides, resulting in impure or incorrect final products after synthesis.
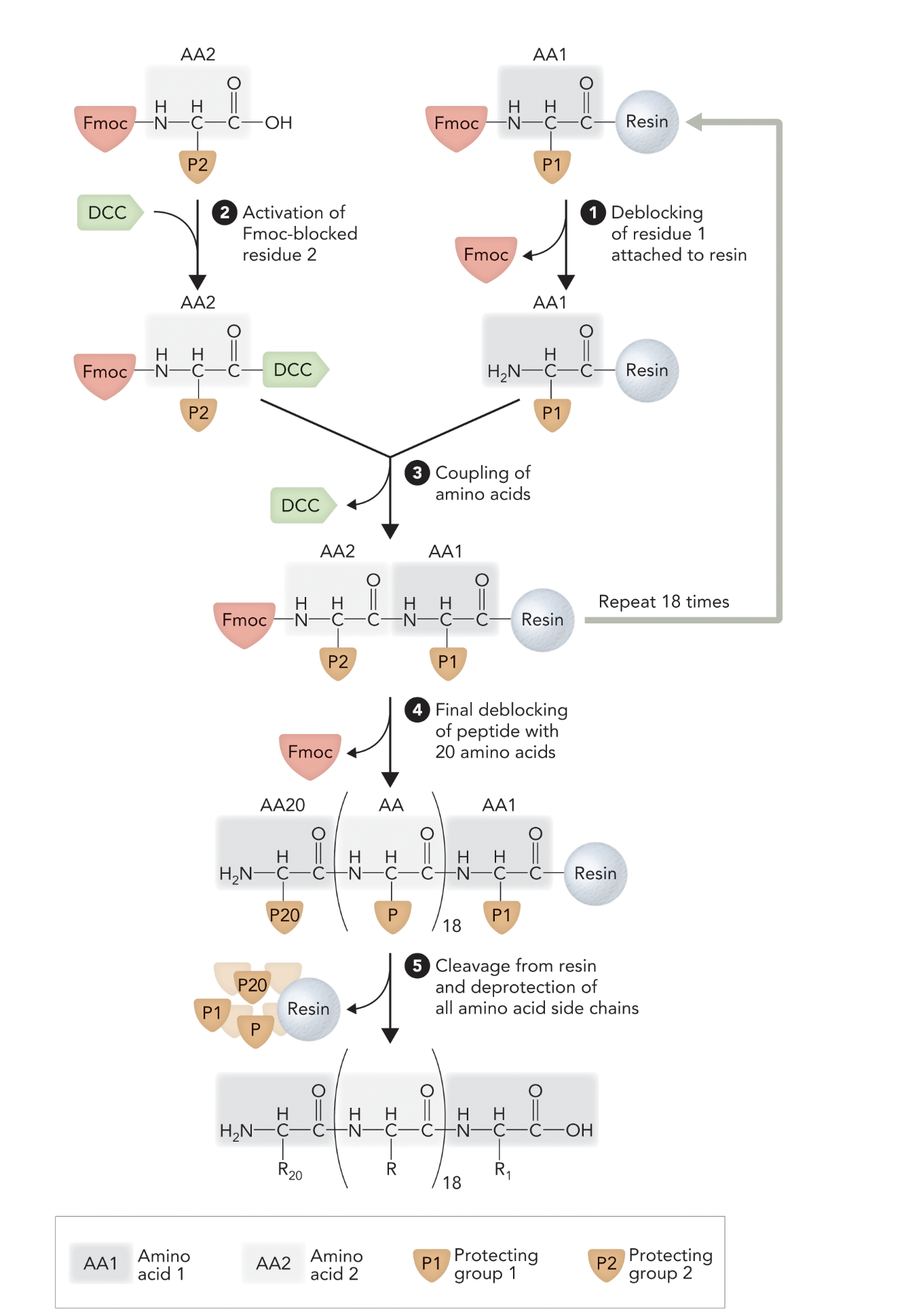
This diagram illustrates the __________ process, a technique used to assemble peptides by sequentially adding amino acids to a growing chain that is anchored to an insoluble resin
Solid-Phase Peptide Synthesis (SPPS)
When the Fmoc protecting group is removed from the first amino acid (AA1) bound to resin, what step of solid-phase peptide synthesis is this?
Step 1
When the second amino acid (AA2) is activated by DCC while still Fmoc-protected, what step of solid-phase peptide synthesis is this?
Step 2
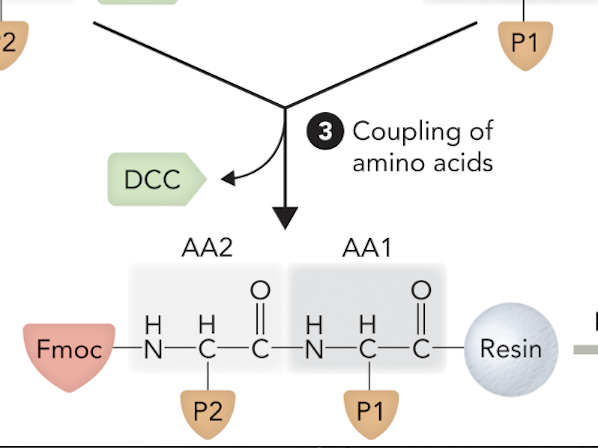
When the two amino acids are joined together via peptide bond formation, what step of solid-phase peptide synthesis is this?
Step 3
When the final Fmoc group is removed from the N-terminal end after all amino acids have been added, what step of solid-phase peptide synthesis is this?
Step 4
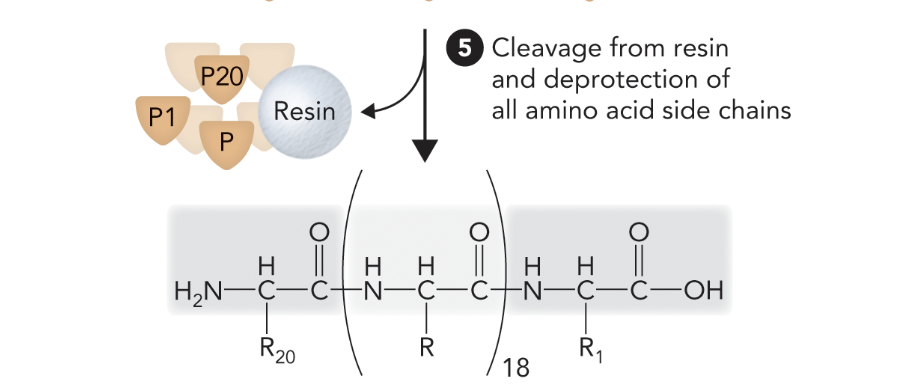
When the peptide is cleaved from the resin and all side chain protecting groups are removed, what step of solid-phase peptide synthesis is this?
Step 5
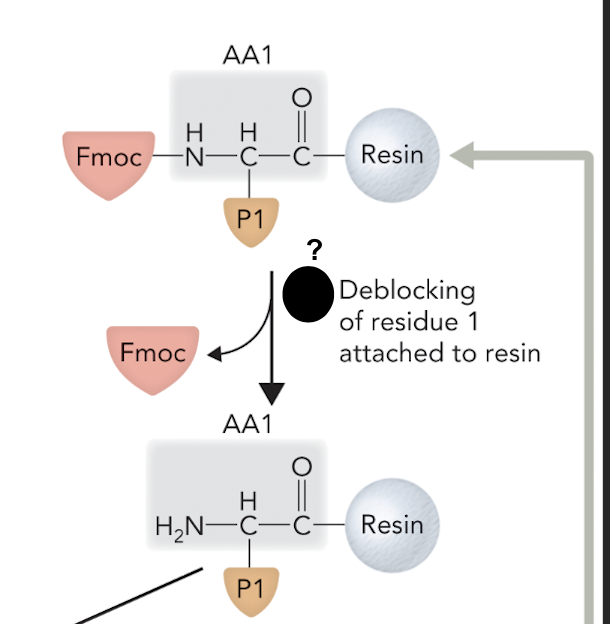
What step is this in SPPS
Step 1
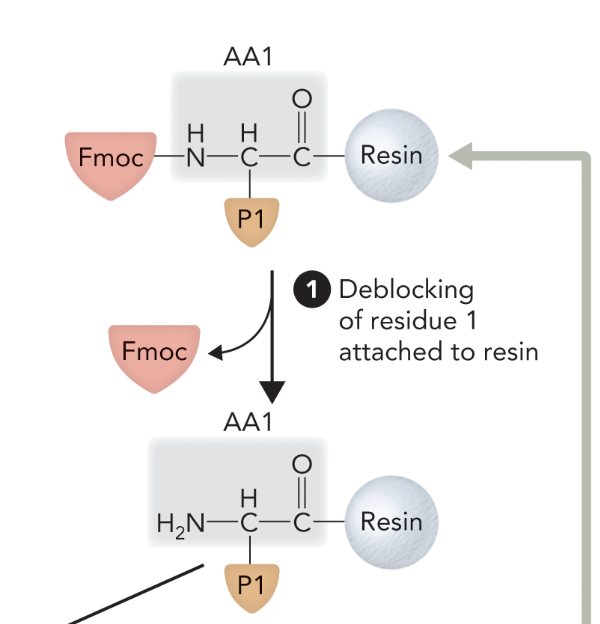
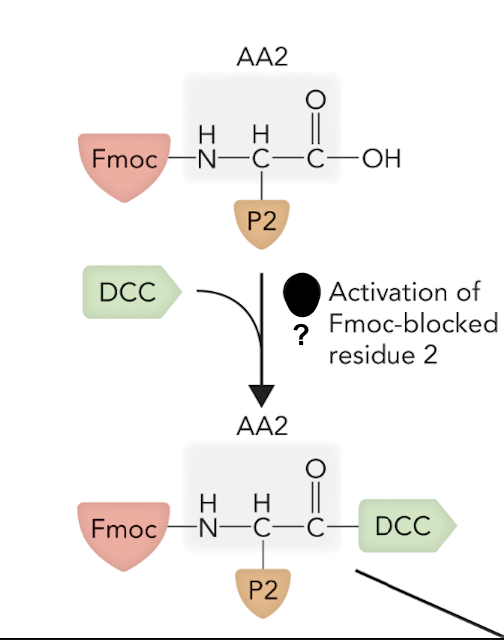
What step is this in SPPS?
Step 2
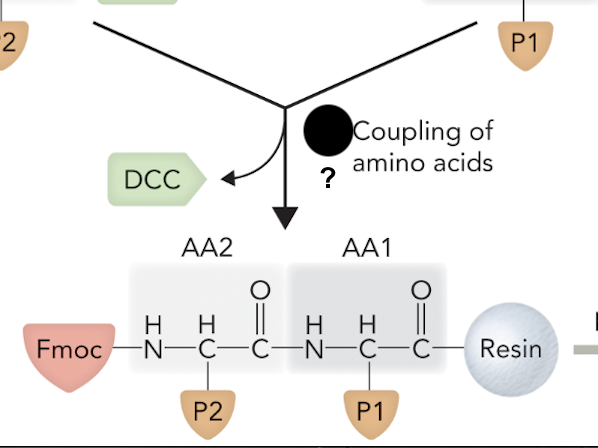
What step is this in SPPS?
Step 3
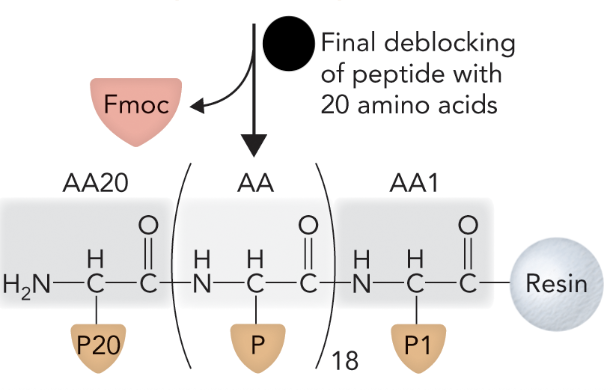
What step is this in SPPS?
Step 4
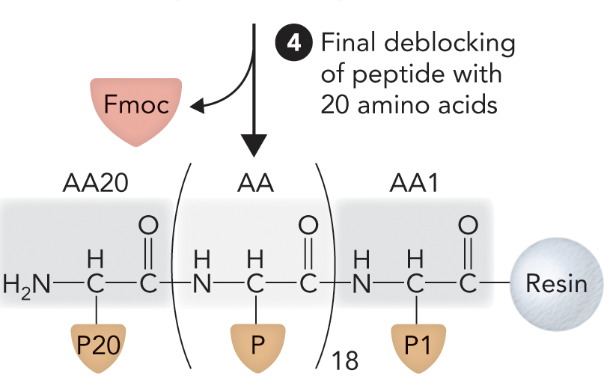
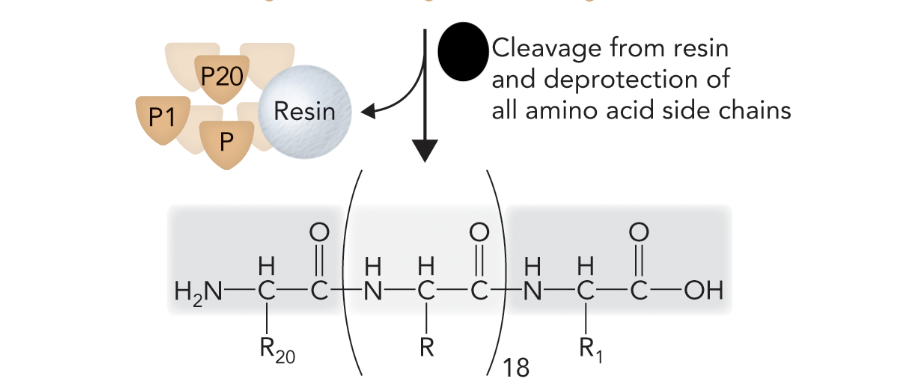
What step is this in SPPS?
Step 5
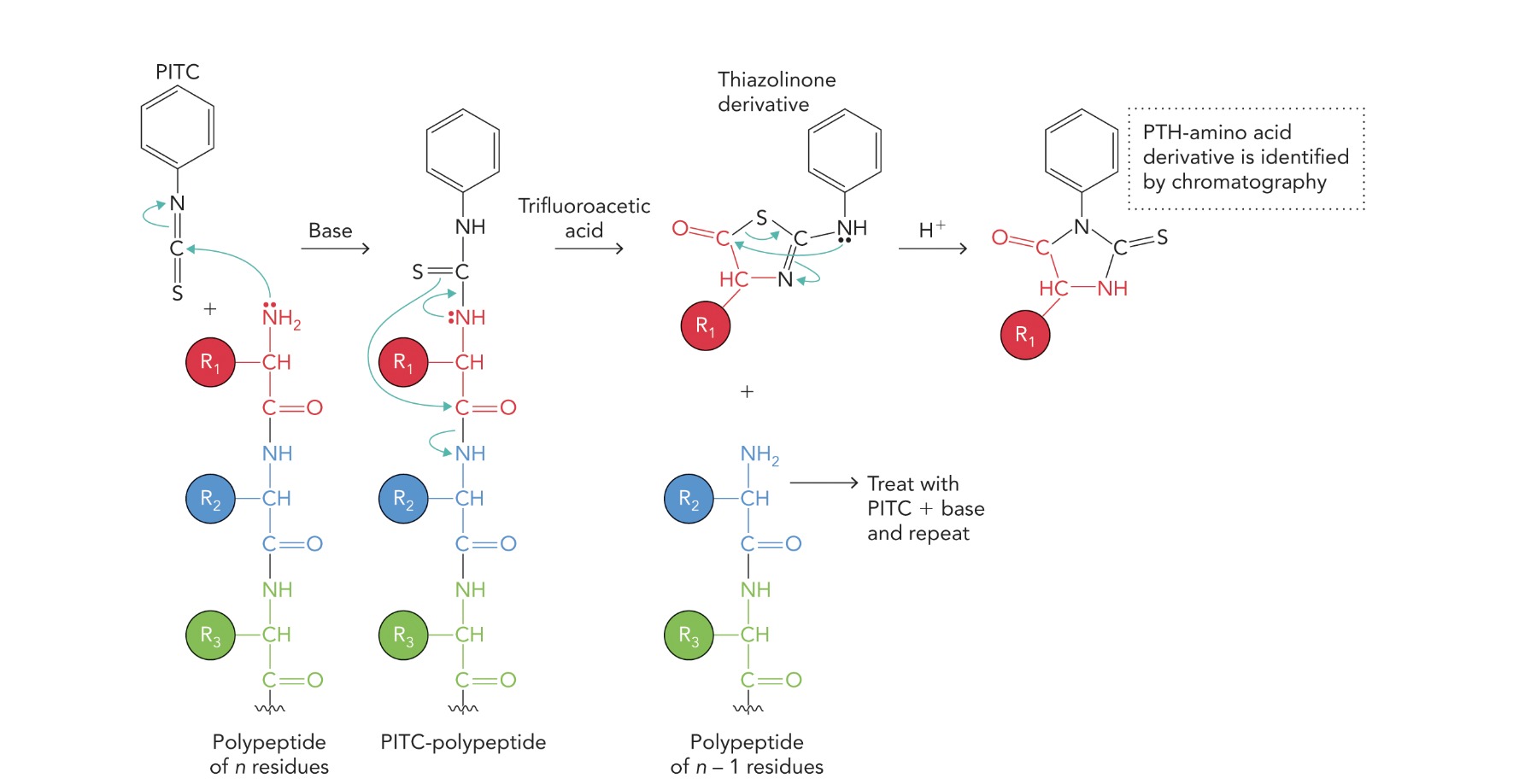
What does the following show?
The diagram illustrates the Edman Degradation process — a stepwise method for sequencing amino acids from the N-terminus of a polypeptide chain.
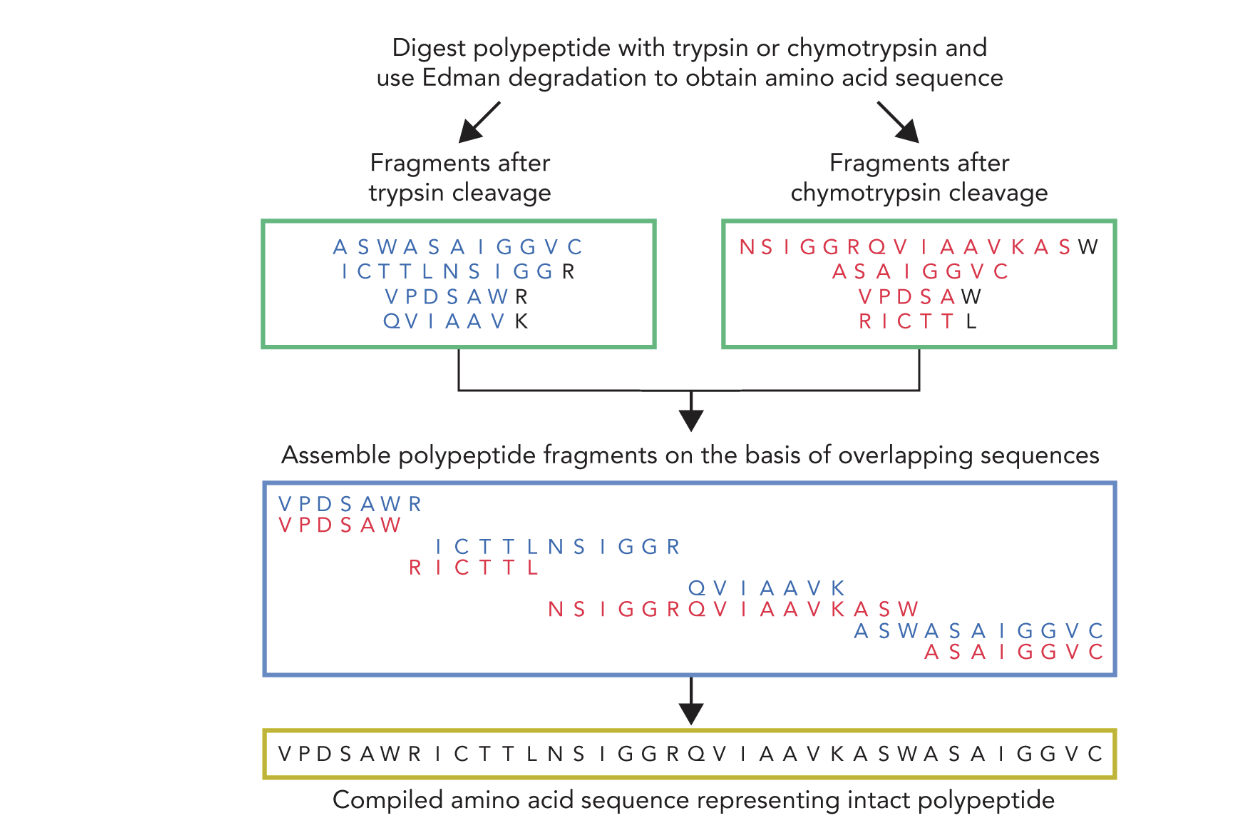
What does the following show?
This diagram shows the strategy for determining the full amino acid sequence of a polypeptide by combining:
🧪 Edman Degradation
— A chemical sequencing method that identifies the N-terminal amino acid of each fragment.
🔪 Protease Digestion
— Using trypsin and chymotrypsin to break the protein into smaller, overlapping fragments that are easier to sequence.
When a polypeptide is digested with trypsin and chymotrypsin to produce short peptide fragments, what step of Edman-based sequencing is this?
Step 1
When each fragment is sequenced individually using Edman degradation, what step of Edman-based sequencing is this?
Step 2
When overlapping regions between fragments from both enzymes are identified, what step of Edman-based sequencing is this?
Step 3
When the full-length polypeptide sequence is assembled by aligning overlapping fragments, what step of Edman-based sequencing is this?
Step 4
When proteins are separated based on charge, size, and abundance, what method is being used?
Gel electrophoresis
What are the two foundational gel electrophoresis techniques used to separate proteins?
SDS-PAGE (based on size) and IEF (based on charge)
When SDS-PAGE and IEF are combined in one process, what technique is being performed?
2-D PAGE (two-dimensional polyacrylamide gel electrophoresis)
Why is 2-D PAGE useful in proteomics?
It enables deep analysis of complex protein samples by separating proteins in two dimensions (charge and size).
When proteins are separated using a polyacrylamide gel matrix and an electric field, what technique is being used?
Polyacrylamide Gel Electrophoresis (PAGE)
What two protein properties does PAGE separate by?
Charge and size
When a gel is placed between two buffer chambers with an applied electric field, which sides are charged?
Cathode = negative (-)
Anode = positive (+)
What provides the electrical current needed for PAGE?
An external power supply connected to the buffer chambers.
What does SDS do to proteins in SDS-PAGE?
SDS (sodium dodecyl sulfate) denatures proteins and imparts a uniform negative charge.
In SDS-PAGE, toward which electrode do proteins migrate?
Proteins migrate toward the anode (+ charge).
In SDS-PAGE, how does protein size affect migration speed?
Proteins migrate at rates inversely proportional to their molecular mass—smaller proteins move faster.
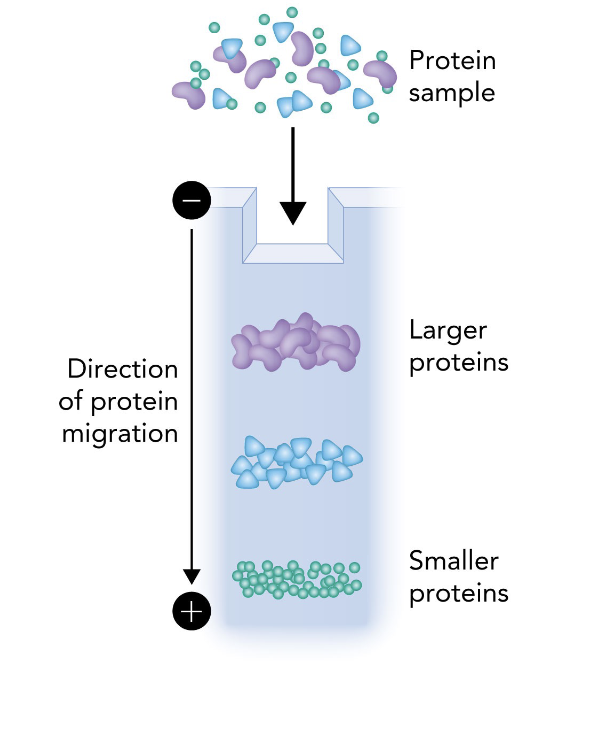
What does the following show?
This image shows the basic setup and principle of SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis), specifically how proteins are separated by size.
A protein sample is loaded into the top of a polyacrylamide gel.
An electric field is applied, with the cathode (-) at the top and the anode (+) at the bottom.
Proteins, coated in SDS (which gives them a uniform negative charge), migrate toward the anode.
Smaller proteins migrate faster through the gel matrix and travel farther.
Larger proteins migrate slower and remain closer to the top.
In SDS-PAGE, which proteins migrate faster through the gel — large or small?
Small proteins migrate faster than large proteins.
What determines the pore size of the polyacrylamide gel in SDS-PAGE?
The acrylamide concentration.
What type of acrylamide gel should be used to resolve large proteins?
A low % acrylamide gel (larger pores).
What type of acrylamide gel should be used to resolve small proteins?
A high % acrylamide gel (smaller pores).
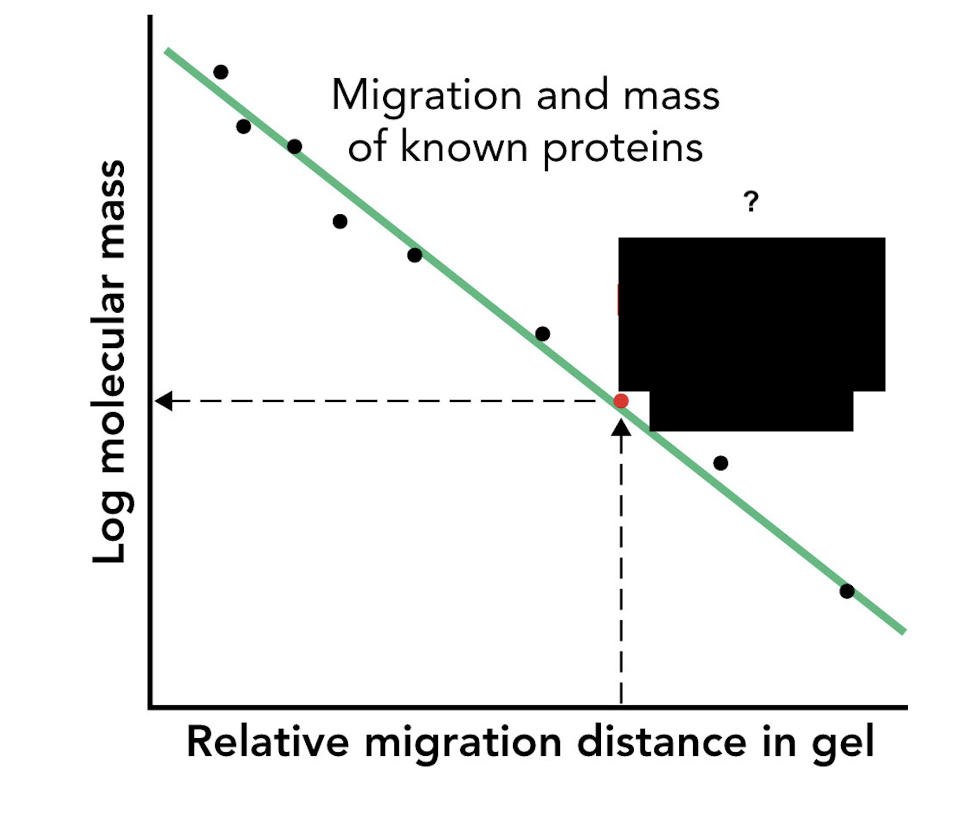
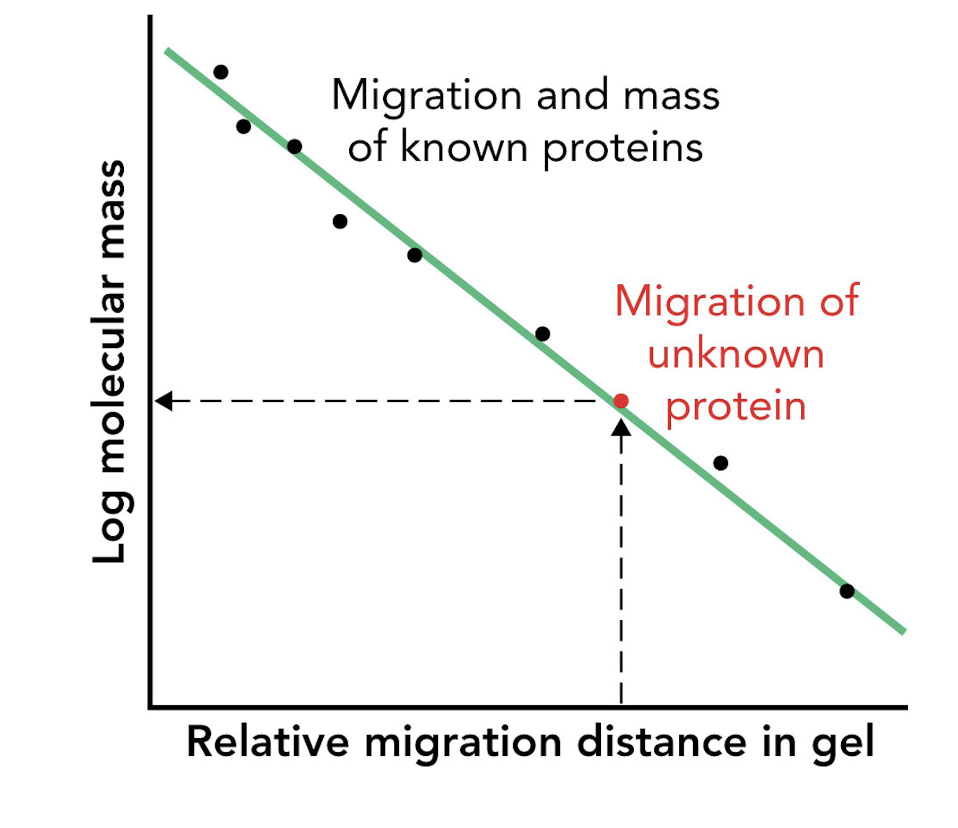
How can the molecular mass of an unknown protein be estimated using SDS-PAGE?
By comparing its migration distance to that of molecular mass markers (protein standards).
What type of graph is used to estimate protein size in SDS-PAGE?
A log(mass) vs. migration distance plot.
What does a log(mass) vs. migration distance plot provide in SDS-PAGE analysis?
A standard curve used to estimate the apparent molecular mass of unknown proteins.
What does Coomassie Brilliant Blue G-250 bind to in proteins?
It binds to basic amino acids, staining proteins in SDS-PAGE gels.
What does staining with Coomassie allow you to visualize in SDS-PAGE?
It allows detection and comparison of protein bands based on size and abundance.
In the SDS-PAGE of a His-tagged protein purified by affinity chromatography, what does the appearance of a ~121-kDa band in elution fractions indicate?
It indicates progressive enrichment of the target protein during purification.
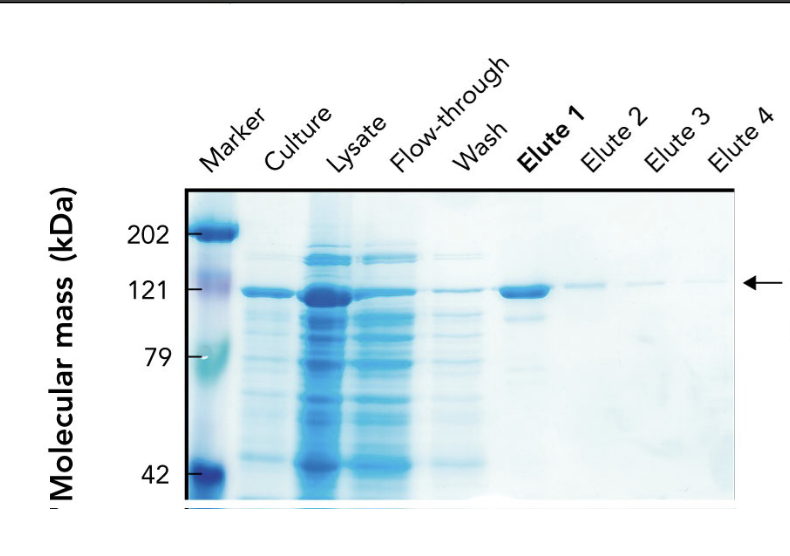
What does the arrow in the SDS-PAGE gel point to?
The target protein (~121 kDa) purified using affinity chromatography.
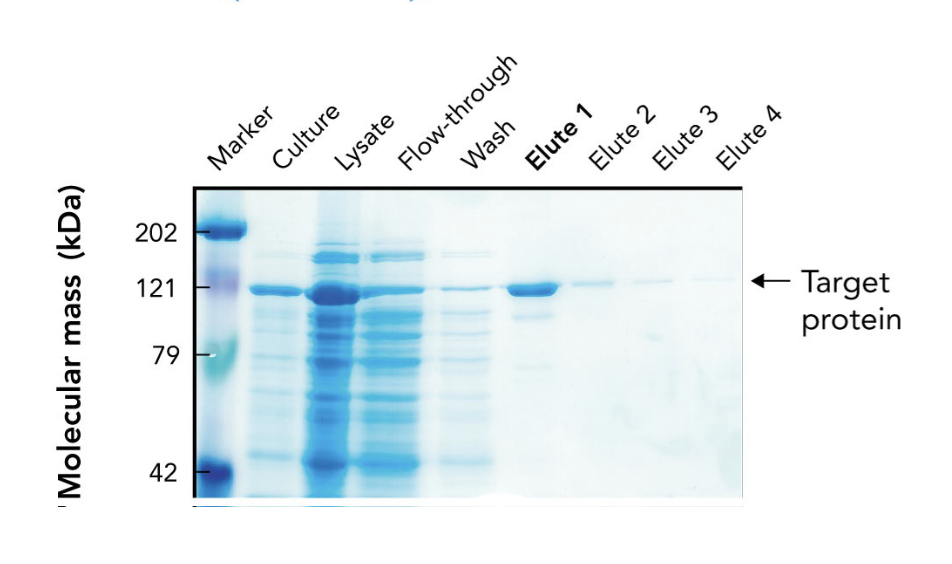
Why is the target protein band lighter than other lanes in the SDS-PAGE gel?
Because it's been purified and enriched, meaning there are fewer total proteins present. It may also stain less if it has fewer basic amino acids for Coomassie to bind.
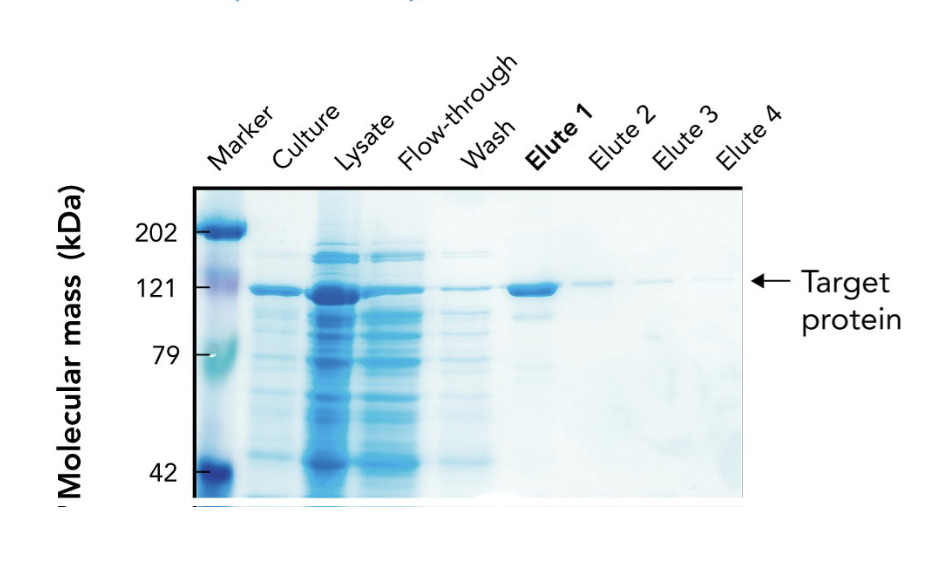
What are "elutes" in protein purification?
Elution fractions — samples collected during elution, where the target protein is released from the purification column using an elution buffer.
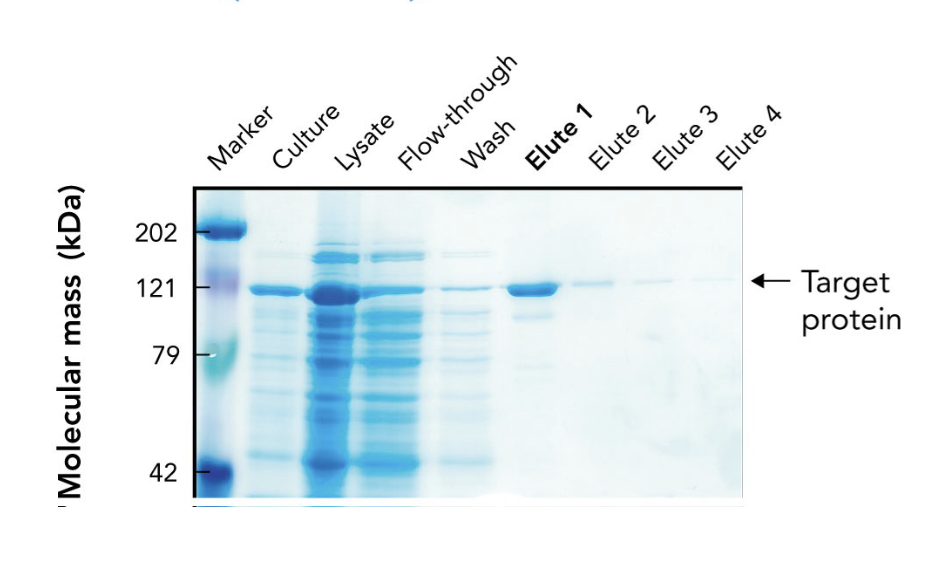
Why are multiple elution fractions (Elute 1–4) collected?
To ensure all the target protein is captured, since it may elute over time in different concentrations.
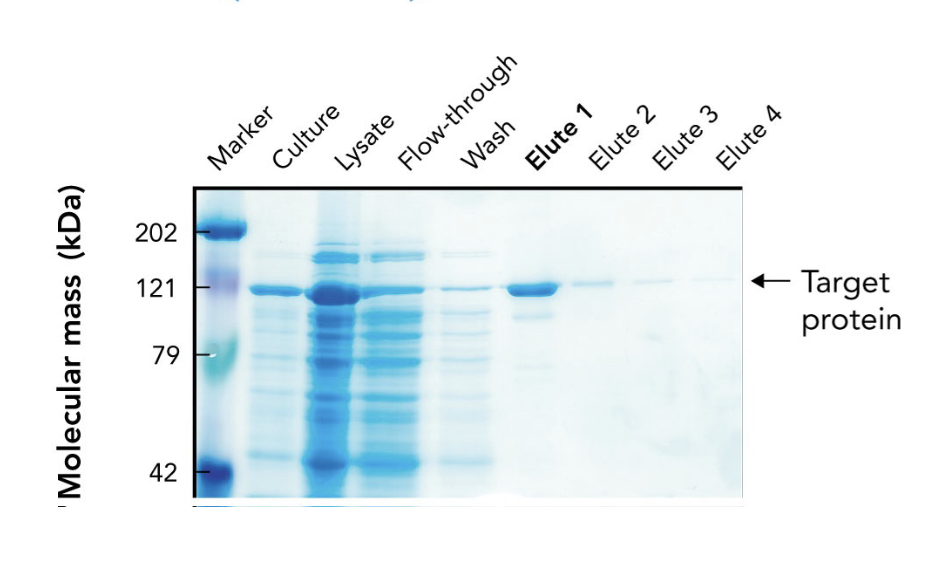
Which elution fraction typically contains the highest concentration of the purified protein?
Elute 1, as seen by the most intense band at the target’s molecular weight.
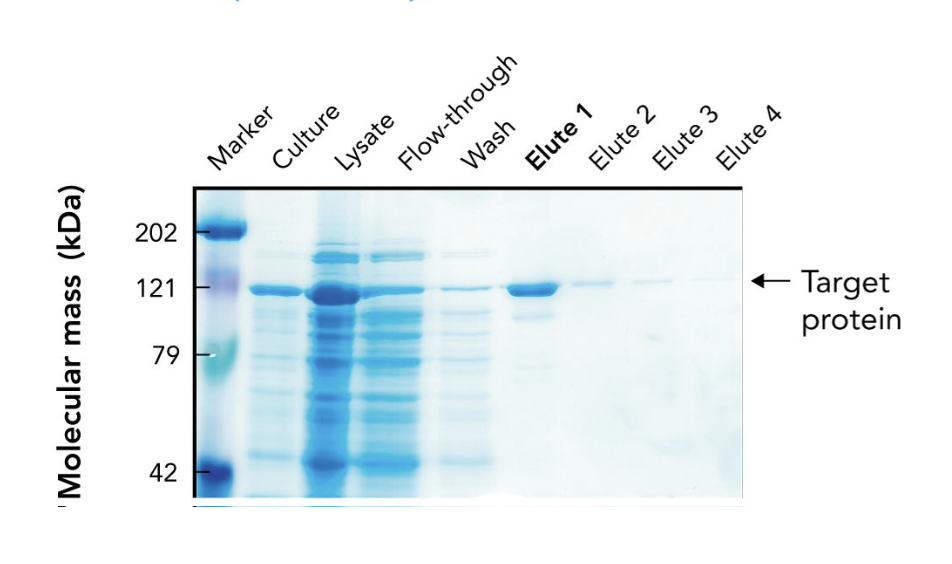
What does the flow-through lane represent in an SDS-PAGE gel during protein purification?
Proteins that did not bind to the affinity column — mostly non-target proteins.
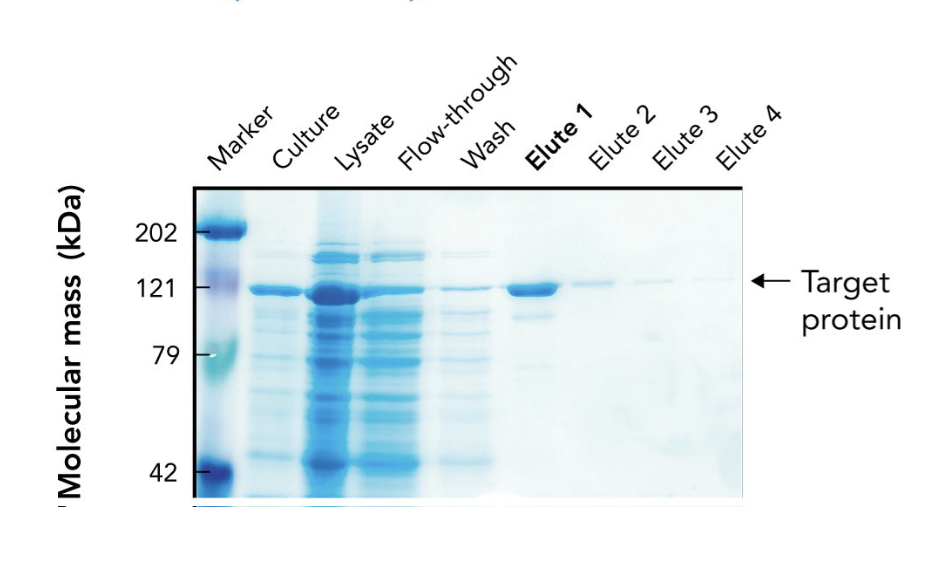
Why is a protein ladder (marker) included in the SDS-PAGE gel?
To estimate the molecular mass of proteins by comparing their migration distances to known standards.
What chemical feature of Coomassie Brilliant Blue contributes to its blue color?
Its aromatic rings contribute to the dye’s color by absorbing visible light.
What type of interaction allows Coomassie dye to bind to proteins?
Electrostatic interactions — the dye’s negatively charged groups (SO₃⁻) bind to positively charged amino acids in proteins (e.g., lysine, arginine).
What functional groups on Coomassie Brilliant Blue provide negative charges for binding proteins?
Sulfonate groups (SO₃⁻)
What property does IEF (Isoelectric Focusing) use to separate proteins?
Their isoelectric point (pI) — the pH at which a protein has no net charge.
What happens to a protein when the pH = pI during IEF?
The protein stops migrating because it has no net charge.
What can be done with gel slices after IEF separation?
They can be cut out, and proteins can be extracted for biochemical assays.
If a protein is protonated or deprotonated, will this tell you if it has a charge?
no
If a protein is deprotonated and its a carboxyl group, will it have a charge? If so what type?
Yes - negative
If a protein is protonated and its a amino group, will it have a charge? If so what type?
Yes - positive
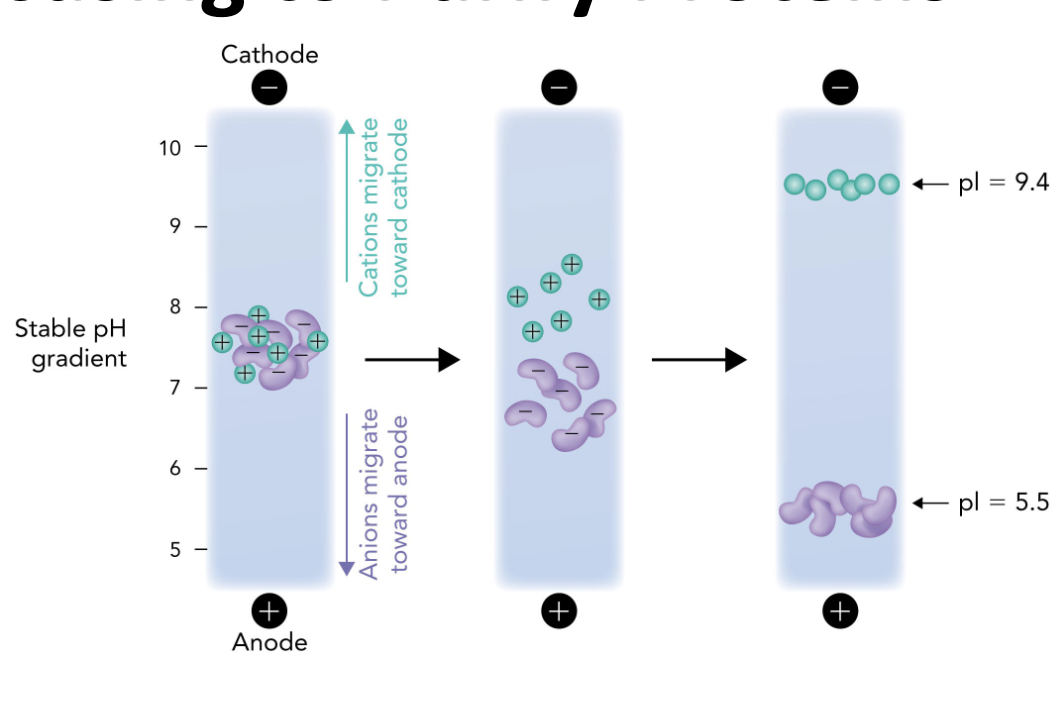
Q: What does isoelectric focusing (IEF) separate proteins based on?
A: Their isoelectric point (pI) — the pH at which a protein has no net charge.
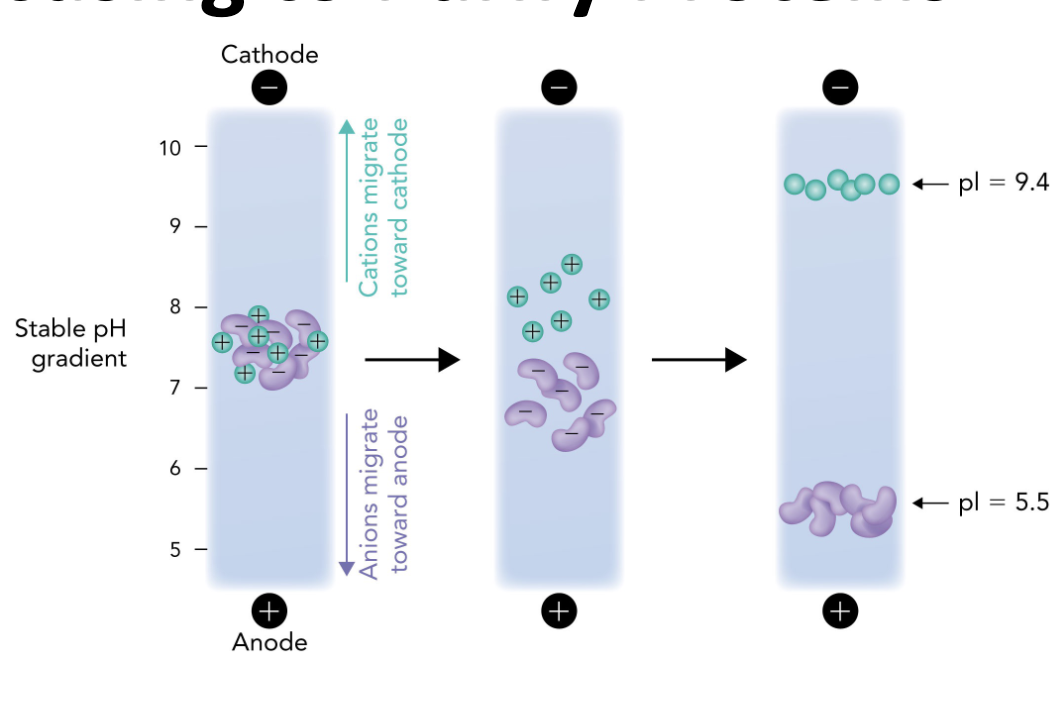
Q: In IEF, what happens when a protein reaches the pH equal to its pI?
A: It stops migrating because it has no net charge (zwitterion)
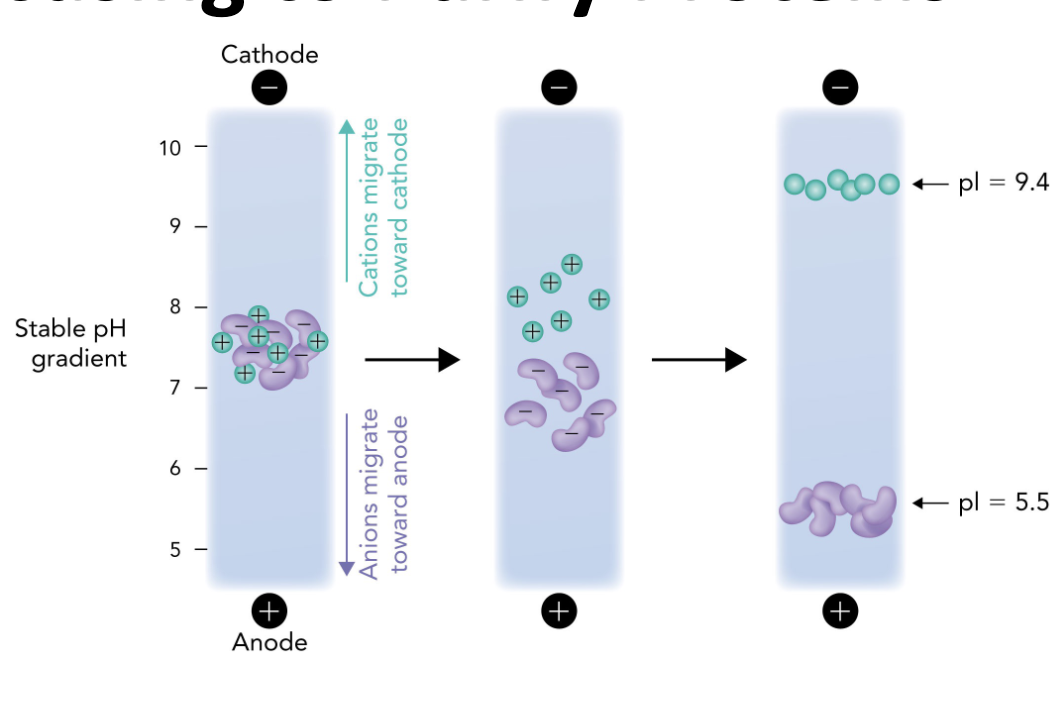
Q: In the IEF gel, which electrode is at the high pH end?
A: The cathode (-) is at the high pH (basic) end.
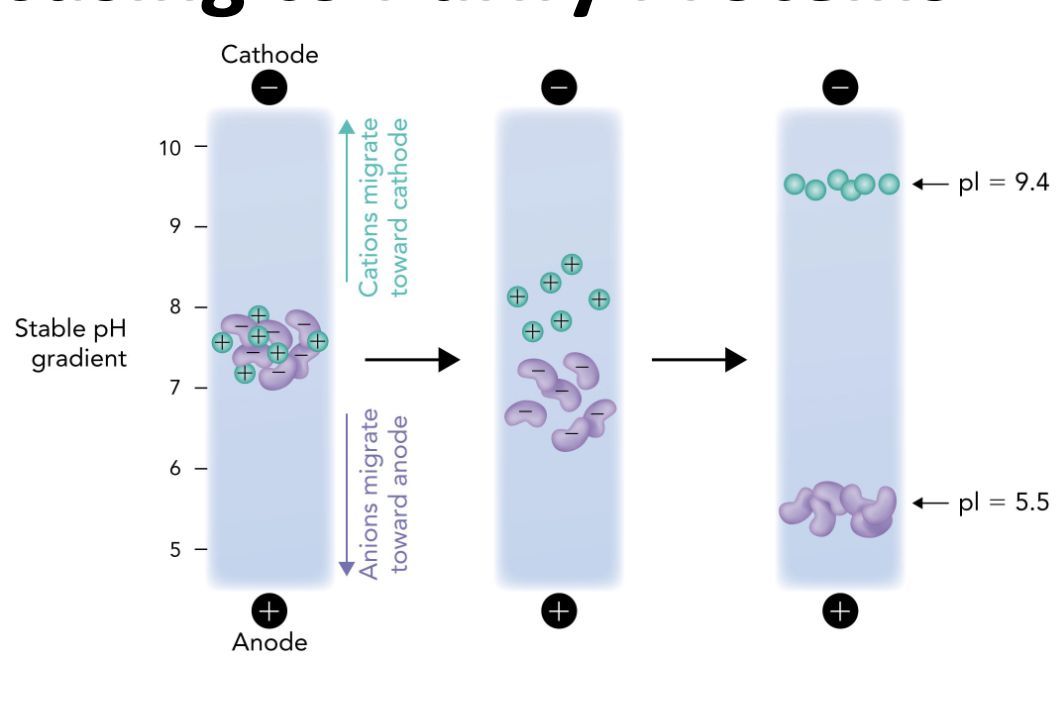
Q: In the IEF gel, which electrode is at the low pH end?
A: The anode (+) is at the low pH (acidic) end.
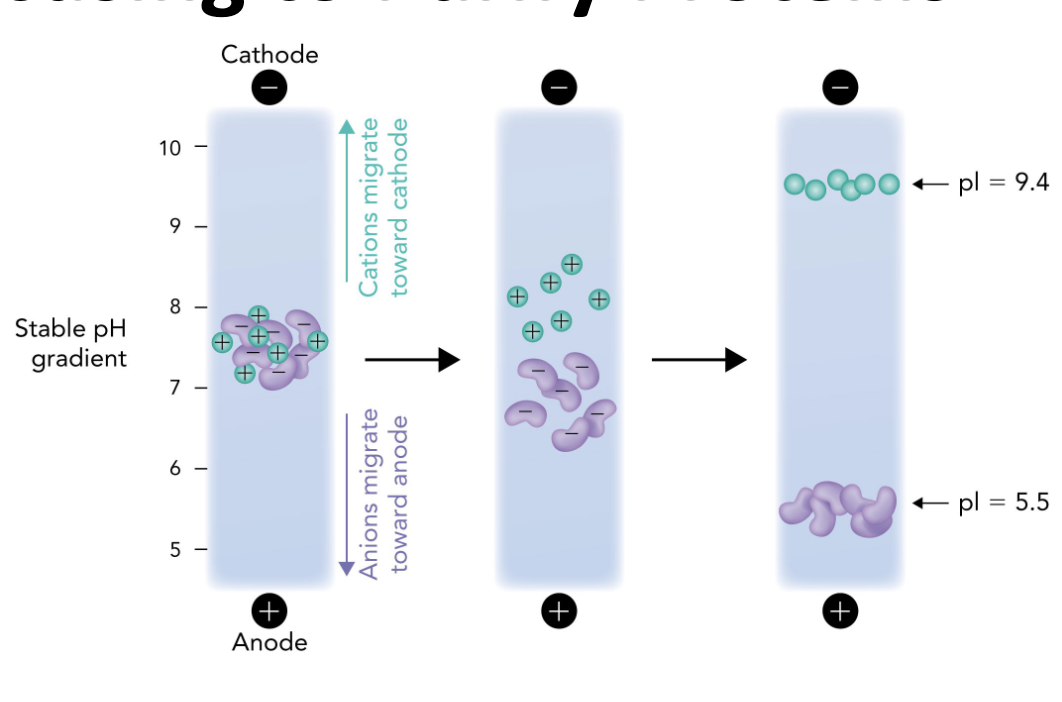
Q: Which direction do positively charged proteins (cations) migrate in IEF?
A: Toward the cathode (-), which is the basic end.
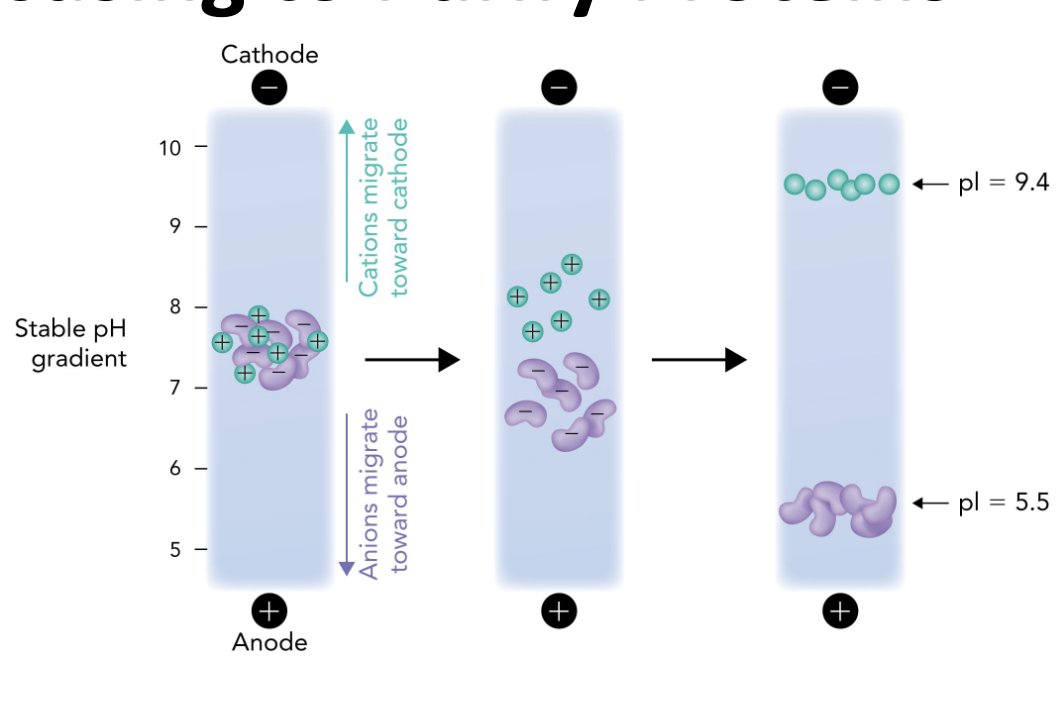
Q: Which direction do negatively charged proteins (anions) migrate in IEF?
A: Toward the anode (+), which is the acidic end.
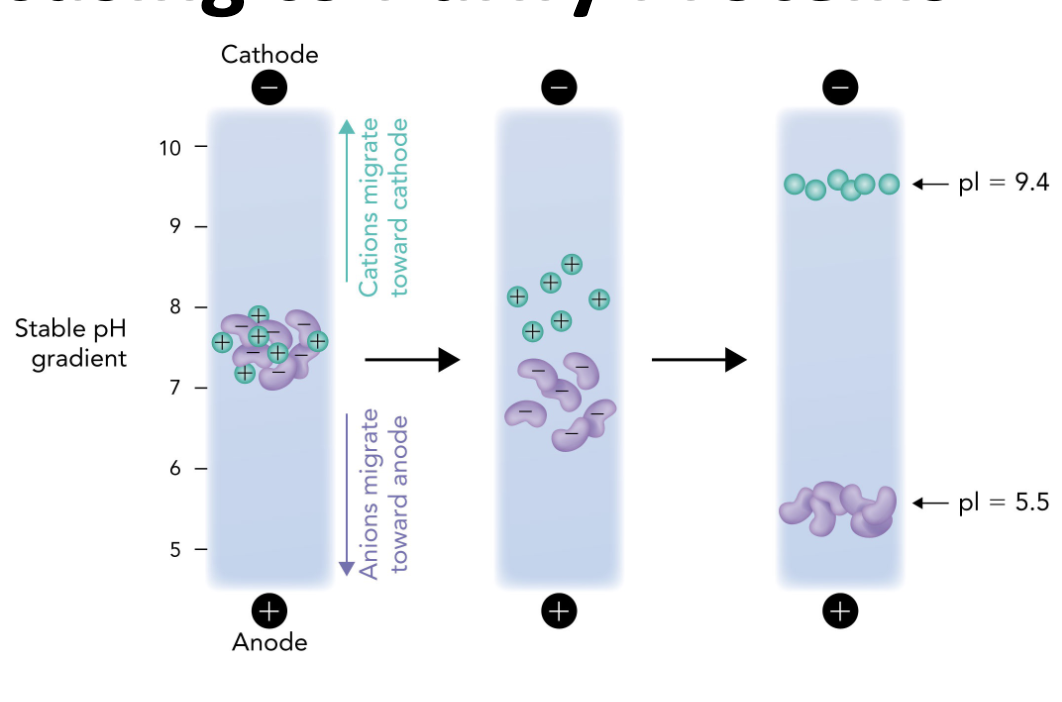
Q: Why do proteins stop moving in an IEF gel?
A: Because they reach a point where the pH = their pI, so they have no net charge and are no longer attracted to either electrode.
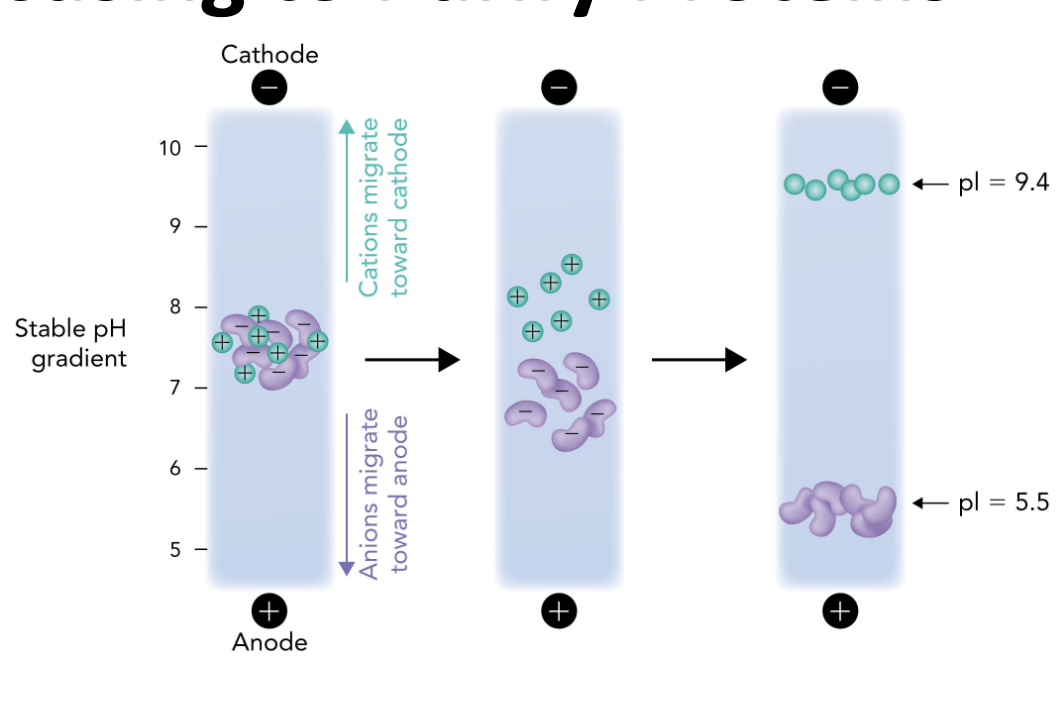
Q: What is the role of the pH gradient in IEF?
A: It allows proteins to migrate to their pI and separate based on charge differences.
Q: What happens when a protein reaches its isoelectric point (pI) during IEF?
A: It becomes a zwitterion with no net charge and stops migrating in the electric field.
A ____ is where there is no net charge of a protein. This is when its reached its isoelectric point
zwitterion
The isoelectric point is? What happens as a result?
The protein has no net charge (it can still be charged) which causes it to stop moving
Q: What two techniques are combined in 2-Dimensional Gel Electrophoresis (2-D PAGE)?
A: Isoelectric focusing (IEF) and SDS-PAGE.
Q: What does the first dimension of 2-D PAGE separate proteins by?
A: Isoelectric point (pI), using IEF.