Antimicrobial Resistance & Drug Discovery
1/48
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
49 Terms
What is antibiotic inactivation or modulation?
Bacteria use enzymes or signaling pathways to DISARM [Degrade] antibiotics, rendering them ineffective
e.g., beta-lactam antibiotics degraded by beta-lactamases
![<p>Bacteria use <strong><mark data-color="red" style="background-color: red; color: inherit">enzymes </mark></strong><mark data-color="red" style="background-color: red; color: inherit">or </mark><strong><mark data-color="red" style="background-color: red; color: inherit">signaling pathways</mark></strong> to <strong><mark data-color="red" style="background-color: red; color: inherit">DISARM [Degrade] antibiotics</mark></strong>, rendering them ineffective</p><p><mark data-color="blue" style="background-color: blue; color: inherit">e.g., beta-lactam antibiotics </mark><strong><mark data-color="blue" style="background-color: blue; color: inherit">degraded </mark></strong><mark data-color="blue" style="background-color: blue; color: inherit">by </mark><strong><mark data-color="blue" style="background-color: blue; color: inherit">beta-lactamases</mark></strong></p>](https://knowt-user-attachments.s3.amazonaws.com/ce79f4fd-ba80-442e-9ee2-80743971e6a1.png)
What is an example of antibiotic inactivation?
Beta-lactam antibiotics are degraded by beta-lactamases, becoming inactive.
What is alteration of target sites in antibiotic resistance?
MUTATIONS in bacterial receptors (e.g., penicillin-binding proteins or ribosomes) PREVENTING antibiotic binding, allowing bacteria to survive.
Reducing residence time of antibiotic results in inactivity.
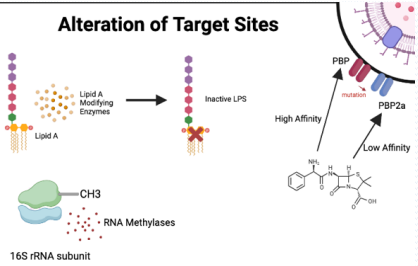
How do mutations in penicillin-binding proteins (PBPs) cause resistance to beta-lactams?
Mutated PBPs block beta-lactam binding, preventing inhibition of transpeptidase and allowing peptidoglycan cross-linking for cell wall formation.
How do mutations in bacterial ribosomes confer resistance?
They prevent ribosome-targeting antibiotics from binding, allowing protein synthesis to continue.
How do mutations disrupt antibiotic binding to receptors?
Mutations (e.g., replacing lysine with phenylalanine) disrupt hydrogen bonds, reducing binding affinity and residency time on the target."
What is outer membrane remodeling in antibiotic resistance?
Gram-negative bacteria modify their outer membrane Porins to block antibiotic entry by:
Mutating porin genes, changing pore size or charge.
Downregulate expression of porins altogether.
preventing access to target sites, common in pathogens like Pseudomonas Aeruginosa .
—
Porin mutation
Bioflim formation expels new debris and matrix content to form a film around site of infection, acting as an additional barrier!
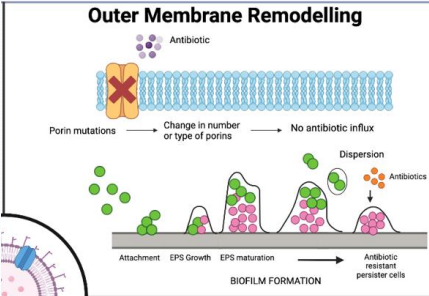
Why is outer membrane remodeling a challenge in gram-negative bacteria?
Their complex membranes (inner, outer, periplasmic space) differ from human cell membranes, and no universal rules predict antibiotic penetration."
What is porin mutation in antibiotic resistance?
Bacteria mutate or remove porins, preventing hydrophilic antibiotics from entering through these membrane channels."
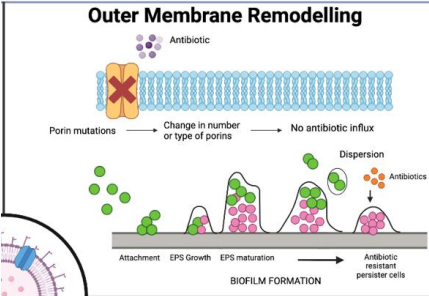
What is biofilm formation in antibiotic resistance?
Bacteria secrete debris and matrix to form protective biofilms, blocking antibiotic penetration, common in catheter-associated infections."
What are efflux pumps in antibiotic resistance?
Machinery that expel antibiotics from the cell, reducing intracellular concentration and promoting resistance."
How many efflux pump types might a bacterium have, and what is their impact?
A bacterium may have 7-8 pump types with hundreds of pumps per cell, non-specifically expelling various antibiotics, lowering their concentration."
What are adjuvants in overcoming antibiotic resistance?
Compounds given with antibiotics to enhance effectiveness by:
Sparing the antibiotic
Enabling its action
Acting as suicide inhibitors.
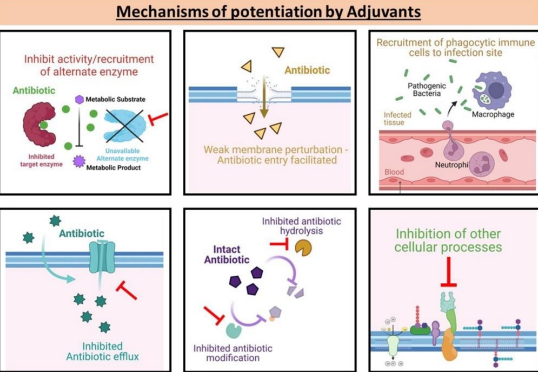
What is an example of adjuvant use in antibiotic therapy?
Beta-lactamase inhibitors paired with beta-lactam antibiotics, covalently binding to beta-lactamases to allow the antibiotic to remain active.
Co amoxiclav, where clavulanic acid acts as a sacrifice protecting amoxiclav.
What challenges exist in developing combination therapies with adjuvants?
Both compounds must reach bacteria simultaneously with matching pharmacokinetics, increasing regulatory hurdles and costs."
What are permeabilizers in overcoming antibiotic resistance?
Compounds that disrupt bacterial membranes to allow antibiotic entry, overcoming barriers like gram-negative membranes.
What is immune system boosting in antibiotic resistance strategies?
Using immunomodulators with antibiotics to enhance the immune system's ability to clear bacteria, reducing reliance on antibiotics."
What are efflux pump inhibitors, and why are they significant?
They block efflux pumps, maintaining high intracellular antibiotic concentrations to kill bacteria, countering a major resistance mechanism.
What are host-directed therapies in antibiotic resistance?
Targeting host processes to make bacterial infection harder to establish, indirectly enhancing antibiotic efficacy.
Instead of killing the bacteria directly (like antibiotics do), this approach modifies how the host (your body) behaves
—
Blocking host receptors or pathways used by bacteria for entry or survival
Example: Inhibiting host actin polymerization to prevent intracellular pathogens from invading cells.Modulating the immune system
Boosting antimicrobial peptide production.
Enhancing macrophage or neutrophil function.
Suppressing excessive inflammation that causes tissue damage (e.g., TNF inhibitors).
Targeting host metabolic pathways
Some pathogens depend on host nutrients or metabolites; manipulating these can starve the bacteria.Autophagy modulation
Encouraging host cells to degrade intracellular bacteria via autophagy.
Name the mnemonics created to memorise WHO Priority Antibiotic Resistance List
CRITICAL:
APE (all Gram neg)
Acinetobacter baumannii
Pseudomonas aeruginosa
Enterobacteriaceae
—
HIGH:
Every Stubborn Helicopter Can Spin Nicely
(1st two are gram positive) Endings match in BOLD
Enterococcus faecium
Staphylococcus aureus
Helicobacter pylori
Campylobacter
Salmonellae spp.
Neisseria gonorrhoeae
—
MEDIUM:
Some Hungry Squirrels
(1st is gram positive)
Streptococcus pneumoniae
Haemophilus influenzae
Shigella spp.
What bacteria are listed in the WHO Priority 1: Critical list for antibiotic resistance?
Gram-negative:
Acinetobacter baumannii
Pseudomonas aeruginosa
Enterobacteriaceae (all carbapenem-resistant, Enterobacteriaceae also 3rd generation cephalosporin-resistant).
What bacteria are listed in the WHO Priority 2: High list for antibiotic resistance?
Gram-negative:
Enterococcus faecium (vancomycin-resistant)
Salmonella spp.
Helicobacter pylori (clarithromycin-resistant)
Campylobacter
[SPIN]
Neisseria gonorrhoeae (fluoroquinolone-resistant, Neisseria also 3rd generation cephalosporin-resistant)
Gram-positive:
Staphylococcus aureus (methicillin-resistant, vancomycin intermediate/resistant). [SPIN]
Every Stubborn Helicopter Can Spin Nicely
What bacteria are listed in the WHO Priority 3: Medium list for antibiotic resistance?
Gram-negative:
Shigella spp. (fluoroquinolone-resistant)
Haemophilus influenzae (ampicillin-resistant).
Gram-positive:
Streptococcus pneumoniae (penicillin-non-susceptible)
Some Happy Squirrels
Name the mnemonics created to memorise Gram Positive & Negative Bacteria list
Gram Positive Cocci: End in COCCI
Lactococci
Staphylococci
Streptococci
Viridians
Enterococci
Gram Negative Cocci:
Neisseria
—
Gram Positive Bacilli: Follow BACILLI spelling
Bacillus
Actinomyces
Clostridium
Corynebacterium
Listeria
Gram Negative Bacilli: All end in ELLA
Salmonella
Shigella
Legionella
Klebsiella
Bordetella
Brucella
Francisella
Pasteurella

What is the average cost of antibiotic development?
£1 billion to £3 billion."
Why is antibiotic development unprofitable?
Low revenue due to short treatment duration / Not as many units can be sold as ‘Short Term Medication’
Competition from generics post-patent expiration
High development costs cause delayed profits beyond 20-year patent life.
What was the Golden Age of antibiotic discovery, and why was it significant?
1940s-1960s
Marked by frequent discoveries from soil microbes
Cheap, safe antibiotics developed rapidly (~9 per year)
Less emphasis on concern about resistance due to continuous new developments.
When did the antibiotic discovery void begin, and why?
Since 1984
No new antibiotic classes developed due to struggles culturing soil bacteria
Failure of high-throughput screening
Antibiotic misuse increasing resistance
How much was invested in antibiotic R&D from 2011-2020 compared to oncology?
Antibiotic R&D remained at $0.2 billion, while oncology R&D grew from $0.8 billion to $6.9 billion."
What did the Wellcome Trust Report (2022) state about antibiotic discovery?
Only 72 drug programs globally are addressing antibiotic discovery, highlighting the limited efforts."
What is Payne's Law in antibiotic development? [Important for MCQs]
Target and cell-based high-throughput screening for antibacterial agents is FIVE times LESS effective than for OTHER drug targets
True across all targets and screening groups.
What was the outcome of 1990s high-throughput screening for antibiotics?
Failure:
Despite ~$100 billion investment, NO NEW antibiotics were developed
As compounds FAILED to PENETRATE bacterial membranes, contributing to the discovery void.
What is the role of CARB-X in antibiotic development?
A collective initiative involving governments and NGOs [Non-Governmental Organization] to fund antibiotic discovery, reporting a £2.8 billion loss for private investors.
What happened to Achaogen, a biotech in antibiotic discovery?
Received FDA approval for a new antibiotic in 2018 but filed for bankruptcy in 2019 due to sales under $100 million against projected $1 billion."
What is phenotypic drug discovery, and why was it effective pre-1990?
Screening compounds against WHOLE bacteria cell to identify hits
Effective because it ensured compounds were ABLE to PENETRATE cells, with target identification done LATER
Target was linked to biological response.
Target is druggable and there is more novelty.
How did antibiotic discovery shift post-1990?
Shifted to high-throughput screening targeting specific bacterial targets after genome decoding
BUT FAILED due to compounds' INABILITY to PENETRATE bacterial membranes.
What are the characteristics of a good antibacterial target?
A good antibacterial target should be:
Present in required bacterial spectrum / Absent in humans
Essential for bacterial growth
Expressed during infection, with known function (e.g., cell wall biosynthesis, membrane integrity, ribosome function).
What is the proposed modified phenotype-based antibiotic discovery approach?
Modified phenotype based antibiotic discovery:
Identify new chemical class by a whole cell screen
Define the spectrum of bacteria that are susceptible by screening AGAINST multi drug resistant isolates [Bacterial strains that are resistant to multiple classes of antibiotics].
Identify the mechanism of actions in vitro [How the compound kills or inhibits the bacteria to understand the target and ensure it’s not a known, resistance-prone mechanism.]
Rationalise observed biological activity using computational techniques [Understand why the compound behaves as it does biologically, guiding optimization of lead compounds and reduce development time.]
Assess the potential for resistance to emerge
Evaluate the in-vivo model [Assess pharmacokinetics (PK), pharmacodynamics (PD), toxicity, and efficacy. Confirm real-world therapeutic potential before moving to humans.]
Identify drug candidate for clinical test evaluation
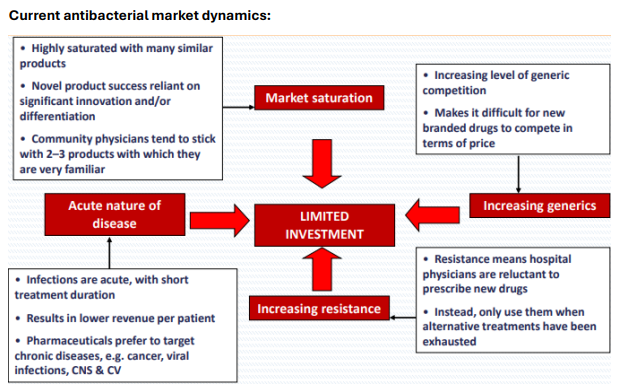
Why is the antibacterial market challenging for new drugs?
Highly saturated with similar products
Physicians prefer familiar drugs
Acute infections yield low revenue
Generics compete on price
What are the contributing factors to limited investment in antibiotics?
Market saturation
Increasing generic competition
Acute (Short Term) nature of infections
Rising resistance discouraging new drug use
What is the 5-year plan to revive antibacterial drug discovery?
Catalytic Phase (Years 1-2): Share knowledge, promote expertise exchange, survey industry data, create chemical repositories
Pilot Phase (Years 3-4): Develop penetration assays, set guidelines, build libraries, test combinations
Implementation Phase (Year 5): Scale libraries, develop nontraditional therapies.
How do combination therapies help in antibiotic discovery?
Target multiple bacterial mechanisms to reduce resistance, increasing available targets, commonly used for tuberculosis.
What are silent operons, and how can they aid antibiotic discovery?
Silent Operons - Inactive bacterial gene systems controlling natural product production
Activating them via chemical messengers or genetic editing could unlock new antibiotics from unculturable bacteria.
What is the Waksman Revival in antibiotic discovery?
Reviving Selman Waksman's culturing techniques to access unculturable bacteria [make up 99% of the total diversity]
As shown by the 2015 discovery of teixobactin, though funding remains a major obstacle.
—
Teixobactin - How you can culture previously unculturable bacteria to produce new antibiotics.
What is teixobactin, and why is it significant?
A new antibiotic class discovered in 2015 from uncultured bacteria
Kills pathogens without resistance, proving unculturable bacteria's potential
Though it failed in clinical trials.
What are prodrugs in antibiotic discovery?
Synthetic compounds activated by bacteria-specific enzymes
Targeting MDR bacteria overexpressing enzymes like beta-lactamases
Reducing selection pressure
—
Prodrug
Early screening for synthetic compounds produced mainly prodrugs suggesting high probability for their discovery.
Prodrugs can enter the cell and are activated by bacteria specific enzymes.
Prodrugs can be selectively targeted against MDR bacteria overexpressing the activating enzyme
Prodrugs can reduce selection pressure and allow opportunities for narrow spectrum antibiotics.
Broad spectrum antibiotics are not always useful as they kill the healthy bacteria as well.
For example, prodrugs which can target bacteria that express a high level of beta lactamases, compared to some wild type bacteria that do not express beta lactamases at all.
How do species-specific compounds reduce antimicrobial resistance?
By specifically targeting a specific species (identified via whole-genome sequencing), it reduces resistance burden on bacteria.
Prevents rapid rise of antimicrobial resistance.
What is the challenge of gram-negative membrane penetration in antibiotic discovery?
Gram-negative bacteria's complex envelope BLOCKS broad-spectrum antibiotic entry
With no predictive penetration rules.
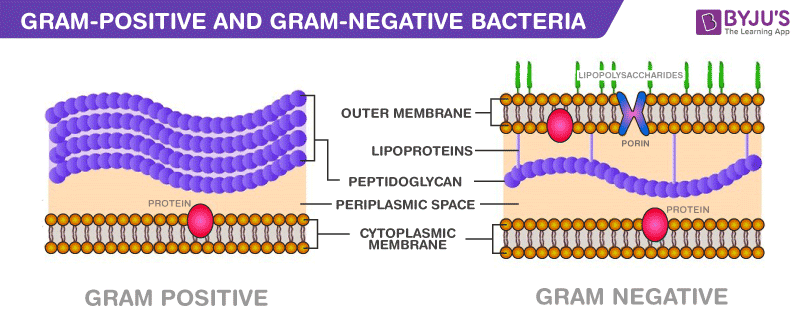
What is the proposed approach to develop rules of penetration for gram-negative bacteria?
Use COMPUTATIONAL techniques: [To rationally design chemical structures likely to penetrate Gram-negative outer membranes.]
To design membrane-active probes [Small molecules or tagged analogs predicted to have penetration]
Synthesize them
Study penetration with microbiology/molecular biology [Experimentally validate the ability of probes to enter Gram-negative bacteria such as mass spectrometry or fluorescent tagging to quantify intracellular accumulation]
To create a 'rule of penetration' for focused libraries.
—
Traditional HTS often failed against Gram-negative bacteria due to poor compound permeability. [Payne’s Law]
The rule of penetration defines chemical features that allow compounds to:
Cross the outer membrane
Avoid efflux pumps
Resist degradation by enzymes like β-lactamases
Enables design of focused chemical libraries with both target-binding and penetration capabilities.
Helps restructure HTS platforms to screen only promising compounds.
Increases number of hits/leads with real antibacterial potential.
Supports rational design by guiding necessary vs. unnecessary features in molecules.
Can revive and improve existing narrow-spectrum antibiotics.
Compounds designed using the rule may be more effective in resistant environments (e.g., biofilms).