BIO230 L3 Prokaryotic Transcriptional Regulation
1/96
Earn XP
Description and Tags
just the names of things
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
97 Terms
transcriptome analyses
provides a signature of cell type
response to extracellular stimuli
3 steps for RNA processing
5’ capping
RNA splicing
3’ polyadenylation
RNA capping (what is it and 3 functions)
addition of a modified guanine nucleotide to the 5’ end of pre-mRNA (3 enzymes involved)
helps in RNA processing and export from the nucleus
important role in translation of mRNAs in the cytosol (ribosomes bind more efficiently to mRNA w/ 5’ cap)
protects mRNA from degradation (5’ exonucleases will otherwise degrade mRNAs by the 5’ end)
RNA splicing
the process of removing introns (non-coding sequences) using an enzyme complex made up of RNA and proteins called the spliceosome
there are multiple ways to splice RNA termed RNA splicing
sites of proper splicing are bound by EJCs
alternative splicing
different or same cells can produce an RNA transcript differently to make different proteins from the same gene
75% of human genes produce multiple proteins which increase the coding potential of genomes
can be regulated
cap-binding complex (CBC)
binds to cap of mRNA to indicate that it is ready
exon
part of mRNA that is left in — mostly known for being a coding sequence
however there are 2 untranslated regions at 5’ and 3’ called noncoding exons
exon junction complexes
serves as a marker for properly sliced RNA
ribosome knocks off EJC as it translates
if it remains on the mRNA when the ribosome reaches the stop codon, then EJC recruits Upf to degrade mRNA
drosophilia sex determination
involves Sex-lethal, Transformer, and Doublesex which all contain regulated splice sites
females experience a short burst of functional Sxl protein
Sxl protein represses splicing of Sxl and Tra, making functional proteins in females
Tra protein activates splicing of Dsx
Dsx represses male gene expression, organism undergoes female development
an example of how alternative splicing can be regulated
Sex-lethal (Sxl)
splicing repressor
produces a nonfunctional protein in male drosophilia
a regulator of splicing in females:
protein functions to block splice sites to create functional Tra protein
Transformer (Tra)
splicing activator
produces a nonfunctional protein in male drosophilia
a regulator of splicing in females:
protein functions to activate the splicing of Dsx
Doublesex (Dsx)
regulates gene expression
normally represses female gene expression
represses male gene expression if splicing regulated by Tra and Tra2 protein
3’ polyadenylation and termination
once RNA Pol II reaches the cleavage and poly A signals in DNA, RNA polymerase transfers protein complexes CstF and CPSF to RNA and terminates transcription
CstF and CPSF recruit additional cleavage factors to cleave RNA
poly A polymerase adds ~200 A nucleotides to the 3’ end of RNA from ATP (not genome encoded)
poly A tail is bound by poly-A binding proteins
CstF and CPSF
carried by the C-terminal domain of RNA polymerase II as it transcribes a gene
when RNA pol II reach the poly A signal sequence in the DNA, CstF and CPSF are transferred to the RNA
CstF is the cleavage stimulating factor: helps stimulate cleavage at the correct site
CPSF provides specificity for recognizing where it should occur
Poly-A polymerase (PAP)
adds ~200 A nucleotides to the 3’ end of RNA from ATP (not genome encoded)
poly-A binding proteins
bind the poly-A tail to aid in
RNA export (required to exit the cell)
translation (important for conformation, interacting with TiF and ribosomes)
mRNA stability (protecting it from degradation by 3’ exonucleases)
tmRNA
acts as both tRNA and mRNA
recruited to the A site of ribosomes processing broken mRNA
carries an alanine amino acid and acts like a tRNA with no anticodon-codon binding
mRNA part is translated by ribosome to create a recognizable tag for degradation
post-translational gene regulation: mRNA stability in prokaryotes
in prokaryotes, exonucleases rapidly degrade most mRNAs
post-translational gene regulation: mRNA stability in eukaryotes
in eukaryotes, mRNAs are more stable and degradation is regulated
2 main mechanisms which both involve gradual poly-A tail shortening like a timer
carried out by deadenylase (an exonuclease) when mRNA reaches the cytoplasm
decapping followed by rapid 5’ to 3’ degradation
continued 3’ to 5’ degradation
both mechanisms can occur on the same mRNA
cytoplasmic poly-A elongation can also occur to stabilize mRNA
proteins can also interfere with poly-A shortening
mRNA stability: transferrin receptor
transferrin receptor imports iron into the cell when iron is low
in iron starvation, mRNA is stabilized by cytosolic aconitase binding the 3’ UTR region (where poly A tail is) to make a hairpin structure
in excess iron, aconitase binds iron and releases mRNA, exposes the 3’ UTR endonucleolytic site, causing rapid degradation of mRNA
aconitase
helps regulate transcription of transferrin receptor by binding to its 3’ UTR in low iron concentration and releasing the mRNA to be destroyed when bound to iron in high iron environments
mRNA stability: deadenylase
competition between mRNA translation and mRNA degradation
deadenylase shortens the poly-A tail binds the 5’ cap like eIFs
when transcription initiation factors are bound to the 5’ cap and poly-A tail, deadenylase cannot bind and vice versa
microRNAs (miRNAs)
non-coding RNAs that base pair with specific mRNAs to regulate mRNA stability
initially double-stranded, has a 5’ cap and poly-A tail, is “cropped” to remove these features
is “diced” to become single stranded and bound with Argonaute
after special processing, it associates with RNA-induced silencing complex (RISC)
if an extensive match is formed, the target mRNA is sliced in half and degraded
RNA-induced silencing complex (RISC)
seeks mRNA with complementary nucleotide sequences. two outcomes of base pairing are possible.
extensive match: target undergoes SLICING and RISC is phosphorylated to un-pair, leading to RAPID DEGRADATION
less extensive match: target undergoes rapid translational REPRESSION, DEADENYLATION, and mostly EVENTUAL DEGRADATION
RNA interference (RNAi)
RNAi destroys double stranded RNA
initiated by dicer protein complex
small interfering RNAs (siRNAs) can interact with Argonaute and RISC proteins follow the miRNA route to destroy double-stranded RNA
siRNAs can also regulate transcription by interacting with Argonaute and the RNA-induced transcriptional silencing (RITS) complex
found in eukaryotes
RNA-induced transcriptional silencing complex (RITS)
interacts with newly transcribed RNA
recruits chromatin modifying enzymes to repress transcription
histone methylation, DNA methylation, transcriptional repression (similar to how chromatin is used in DNA to suppress genes)
Argonaute
a protein of RISC that plays a critical role in base-pairing miRNA with mRNA
CRISPR-Cas immunity
an immune reaction in prokaryotes
short fragments of viral DNA integrate into the CRISPR region of the genome and become templates to produce crRNAs
viral DNAs complementary to CRISPR regions are directed for degradation by Cas proteins
similarly to Argonaute: use of small single-stranded RNA
CRISPR
a region of the prokaryotic genome where viral DNA is collected to become crRNAs
viral DNAs complementary to CRISPR regions are degraded
CRISPR-associated proteins (Cas)
directs viral DNA complementary to CRISPR regions to degradation
Lac repressor
binds to the lac operator to block RNA polymerase through competitive binding
unbound when lactose (ligand) concentration is high
example of negative regulation (bound repressor prevents transcription)
catabolic activator protein (CAP)
binds to the CAP binding site to recruit RNA polymerase
bound when cAMP (ligand) concentration is high due to low glucose
example of positive regulation (bound activator promotes transcription)
Trp repressor
binds to the Trp operon to block RNA polymerase through competitive binding
bound when tryptophan (ligand) concentration is high
contains helix-turn-helix DNA binding motif
example of negative regulation (bound repressor prevents transcription)
helix-turn-helix
DNA binding motif where protein regulator binds in the major groove of the DNA double helix
negative regulation
competition between RNA polymerase and repressor protein for promoter binding
positive regulation
activator protein recruits RNA polymerase to the promoter to activate transcription
gene regulatory elements are typically found (prokaryotes, eukaryotes)
prokaryotes
close to the transcriptional start site
eukaryotes
far upstream of gene
downstream of gene
within gene (introns)
NtrC protein
transcriptional activator located far from promoter
DNA looping allows NtrC to directly interact with RNA polymerase to activate transcription from a distance
bacteriophage lambda
virus that infects bacterial cells
example of positive and negative regulatory mechanisms work to regulate the lifestyles of bacteriophage lambda
two gene regulatory proteins are responsible for initiating this switch and repress each other’s synthesis
lambda repressor protein (cI)
represses lysis and activates prophage in bacteriophage lambda
occupies the shared operator, blocks synthesis of Cro
activates its own synthesis by recruiting RNA polymerase, most bacteriophage DNA not transcribed
positive feedback loop
Cro
represses prophage and allows lysis in bacteriophage lambda
occupies the shared operator, blocks synthesis of lambda repressor
most bacteriophage DNA extensively transcribed
DNA replicated, packaged, new bacteriophage released by host cell lysis
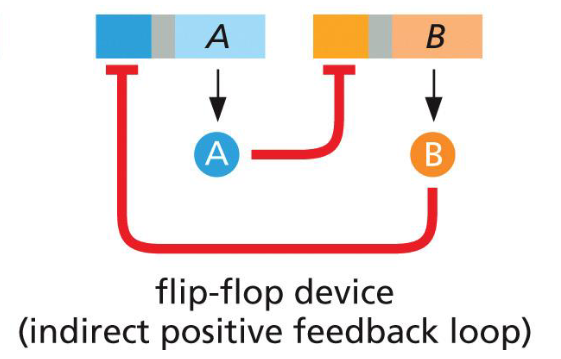
flip-flop device (indirect positive feedback loop)
conditions to trigger the switch between prophage and lysis in lambda bacteriophage
host response to DNA damage — switch to lytic state
inactivates repressor
good growth conditions — maintains prophage state
lambda repressor turns off Cro and activates itself in a positive feedback loop
positive feedback loop (uses)
e.g. lambda repressor protein
can be used to create cell memory — like staying differentiated in cell division
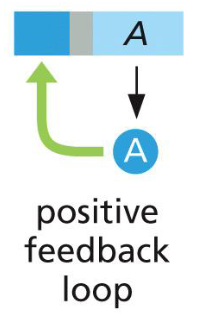
negative feedback loop
more A turns off itself

flip flop device (indirect positive feedback loop)
e.g. Cro/repressor switch
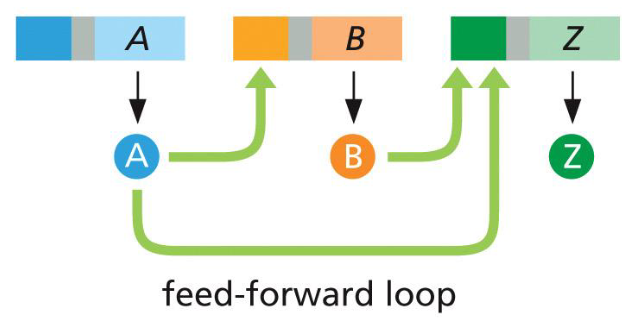
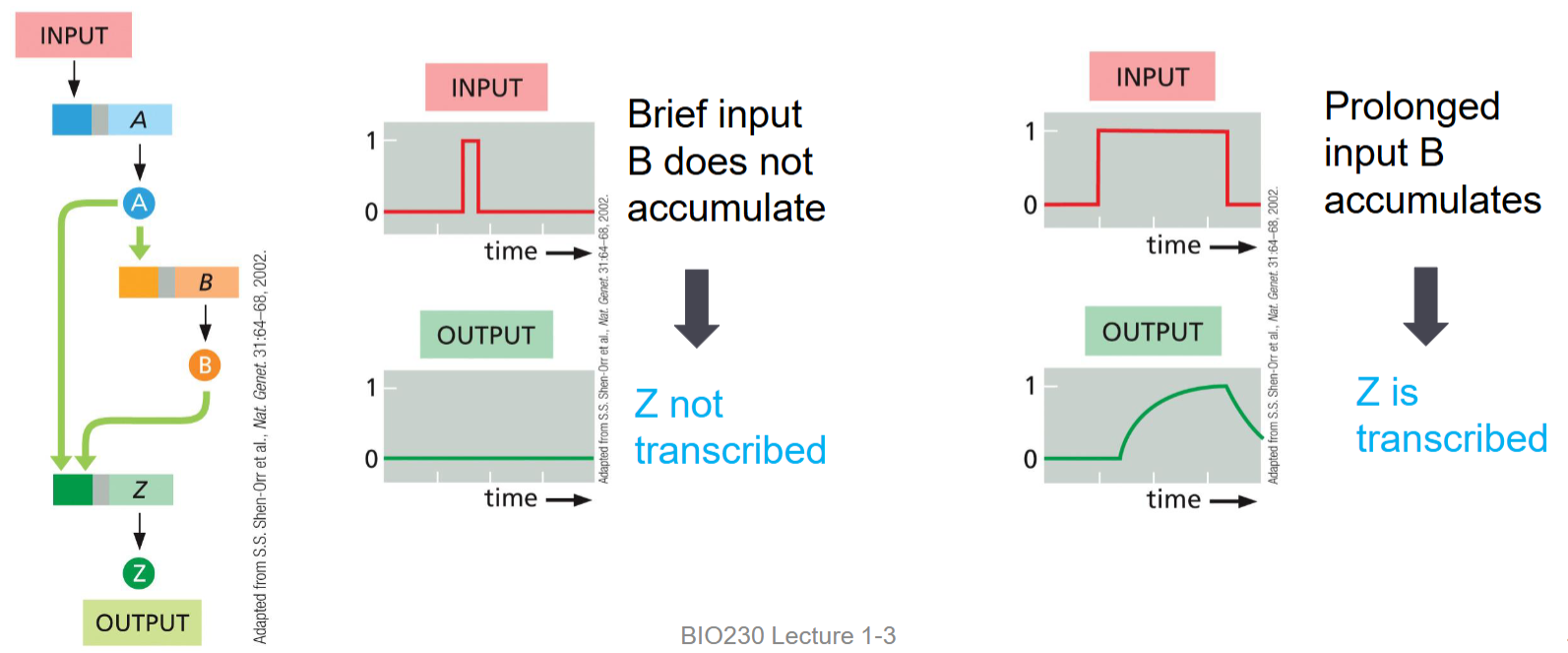
feed-forward loop (uses)
two gene products required to regulate a third
can measure the duration of a signal
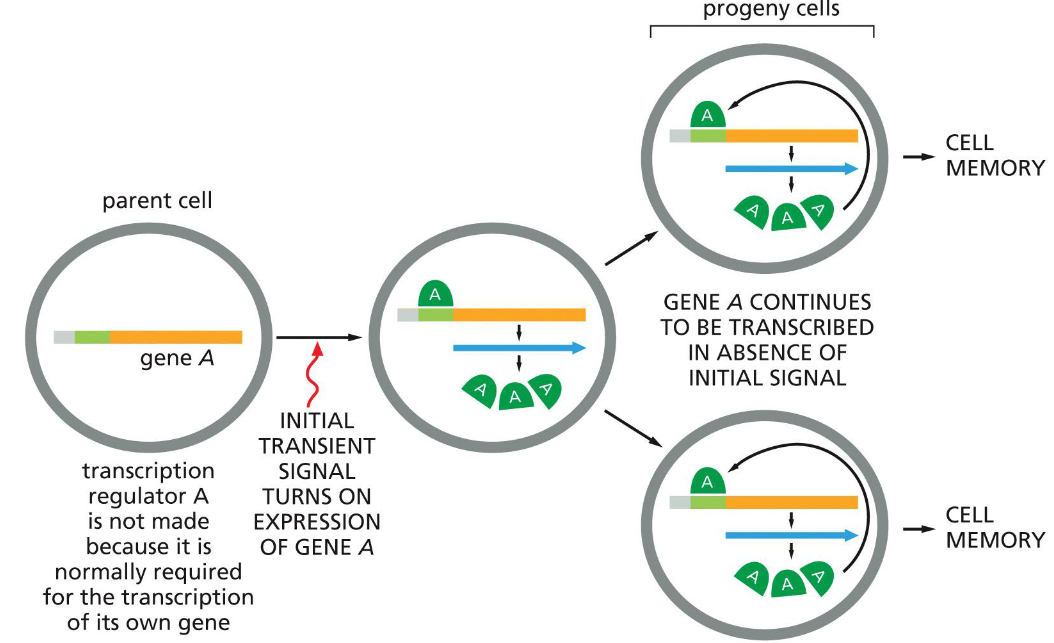
how positive feedback loops can be used to create cell memory
proteins remain in cell after division
how feed forward loops can measure the duration of a signal
relies on accumulation of protein
the repressilator
simple gene oscillator using a delayed negative feedback circuit
however amplitude increased over time due to bacterial growth
an example of synthetic biology
circadian gene regulation in drosophila
uses delayed negative feedback loop to regulate sleep cycle
transcription attenuation
RNA adopts a structure that interferes with RNA polymerase causing a premature termination of transcription
regulatory proteins can bind to RNA and interfere with attenuation
prokaryotes, plants, some fungi use riboswitches to regulate gene expression
riboswitches
short RNA sequences that change conformation when bound by a small molecule
prokaryotes, plants, and some fungi use riboswitches to regulate gene expression
eg prokaryotic riboswitch that regulates purine biosynthesis
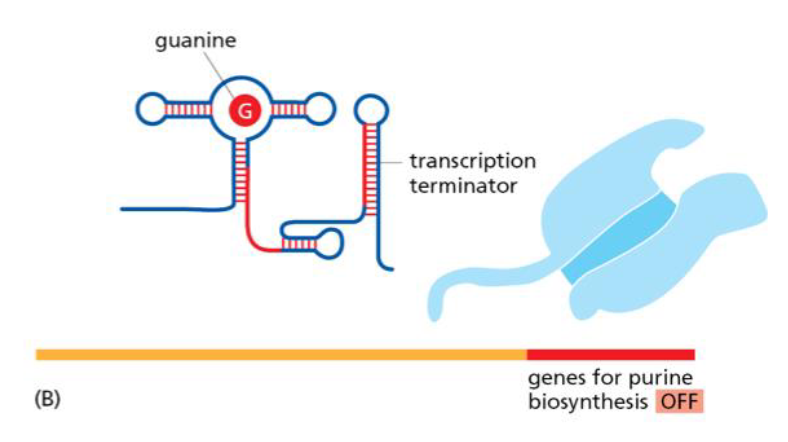
prokaryotic riboswitch that regulates purine biosynthesis
low guanine levels activate transcription of purine biosynthetic genes
high guanine levels cause guanine to bind to riboswitch and RNA polymerase to terminate transcription of purine biosynthetic genes
TFIID
general transcription factor
recognizes TATA box and other DNA sequences (other consensus sequences) near the transcription start point for initiation
bends DNA to highlight it for RNA polymerase recognition
TFIIH
general transcription factor
unwinds DNA at the transcription start point
phosphorylates Ser5 of the RNA polymerase C-terminal domain (CTD)
releases RNA polymerase from the promoter to initiate transcription
RNA polymerase II
transcribes protein coding genes in eukaryotes
general transcription factors
TFIID, TFIIB, TFIIF, TFIIE, TFIIH for eukaryotes
sigma factor for prokaryotes
difference between eukaryotic genomes and prokaryotic genomes
eukaryotic genomes
lack operons
1 gene, 1 RNA molecule, 1 protein
packaged into chromatin (additional mode of regulation)
prokaryotic genomes
operons
multiple genes encoded on 1 RNA molecule
mediator
acts as an intermediate between many regulatory proteins and RNA polymerase in eukaryotes
coactivators and corepressors
only bind to proteins that are already bound to DNA, do not directly bind to DNA
DNA binding domain (DBD)
a module of eukaryotic activator protein that recognizes specific DNA sequence
very specific about binding
activation domain (AD)
a module of eukaryotic activator protein that accelerates frequency/rate of transcription
tends to be general
how activator proteins activate transcription
attract, position, and/or modify general transcription factors, mediator, RNA polymerase II
by either directly interacting with them, or indirectly modifying chromatin structure
activator proteins can bind directly to transcriptional machinery or the mediator and attract them to promoters
nucleosomes
the basic structure of eukaryotic chromatin
DNA wound around a histone octamer with 2 sets of H2A, H2B, H3, H4 creates a compact chromatin fibre that can be unwound or altered to increase promoter accessibility
4 major ways activator proteins can alter chromatin
chromatin remodeling complex:
nucleosome sliding
histone removal
histone replacement
histone-modifying enzyme:
specific pattern of histone modification