Week 5 - Eukaryotic Chromosome Abnormalities and Molecular Organization
1/40
Earn XP
Description and Tags
Chapter 10
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
41 Terms
How do chromosome number and shape vary among species?
Species-specific genome content, chromosome number, size, and shape.
Closely related species tend to have similar chromosome numbers.
Each chromosome pair in a diploid genome has distinct characteristics.
What did Thomas and Christoph Cremer discover about chromosomes in interphase?
Chromosomes occupy specific regions called chromosome territories.
They do not stray from their territory until mitosis occurs.
Each nucleus has a unique arrangement of chromosome territories.
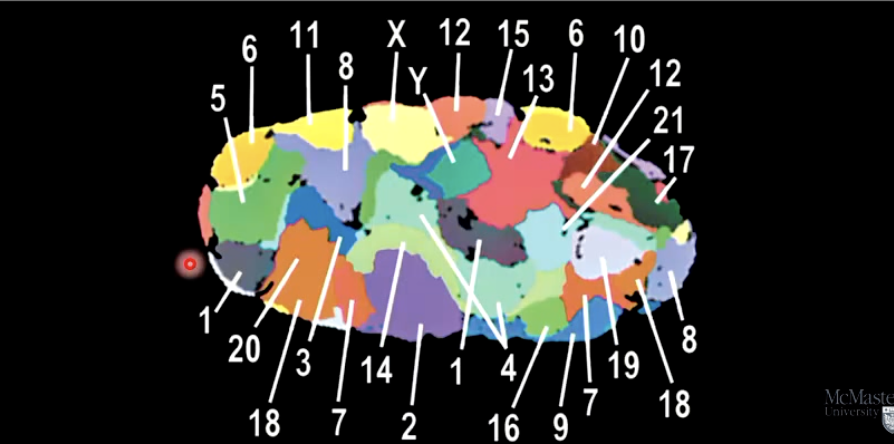
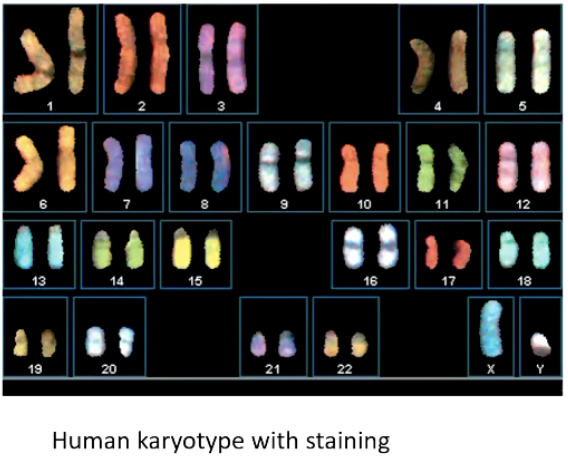
What is a karyotype, and what is it used for?
An organized display of chromosomes, captured using microscopy.
Helps identify abnormal chromosome numbers or structures.
Arranged in descending size order, autosomes first.
Staining techniques can highlight specific chromosomes.
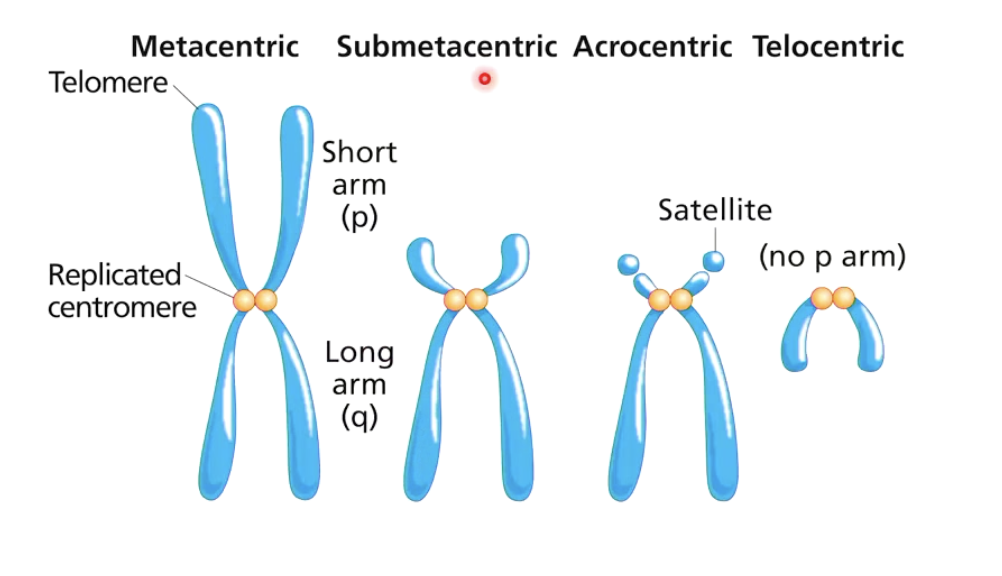
What is the centromere, and the p and q arms of a chromosome?
Centromere: Divides chromosome into two arms.
p arm: Short arm of the chromosome.
q arm: Long arm of the chromosome.
What is FISH, and how is it used?
Fluorescence in Situ Hybridization
Uses fluorescent molecular probes to detect specific DNA sequences.
Probes emit fluorescence at different wavelengths.
Helps identify individual chromosomes within a cell.
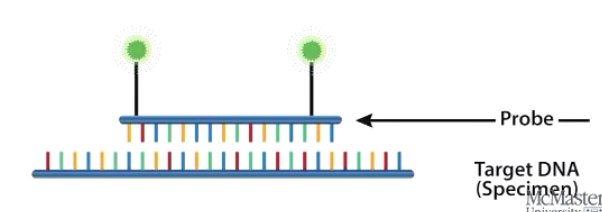

What is the purpose of chromosome banding techniques?
Older method used before FISH.
Identifies chromosomes based on size, shape, and banding patterns when dyes and stains are used.
Process:
Stop cell cycle in metaphase.
Apply stain/dye to chromosomes.
Burst the cell, isolate chromosomes, and photograph them.
May require multiple attempts for clear images.
What is G-banding, and how does it work?
The standard banding technique for human chromosomes.
Uses Giemsa stain to create a unique banding pattern.
Allows for identification of major and minor chromosomal regions
How are chromosome bands labeled?
Uses letters and numbers to mark major and minor band regions.
Numbering starts at centromere and moves outward along each arm.
Each alternating light and dark band is labeled in a hierarchical numbering system.
Example: 5q2.3.1
5 = Chromosome number.
q = Long arm.
2 = Region 2 on the 5 q arm.
3.1 = Specific dark band location.
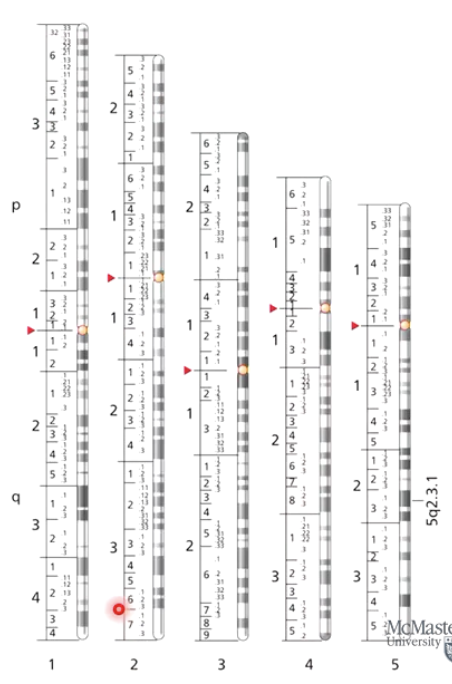
What is the difference between euchromatin and heterochromatin?
Euchromatin: Light bands, less compact, higher gene expression.
Heterochromatin: Dark bands, more compact, lower gene expression.
Who first discovered chromosome banding, and in what organism?
Discovered by Édouard-Gérard Balbiani in 1881.
Found in polytene chromosomes of dipteran flies (e.g., Drosophila).
What are polytene chromosomes, and where are they found?
Found in salivary glands of fly larvae.
Formed by repeated DNA replication without cell division, creating giant chromosomes.
Easily observed under a microscope and produce distinct bands when stained.
How have polytene chromosomes been used in genetics?
Help map genes.
Identify various mutations.
Study chromosome structure and function.
Picture shows how the region in chromosome is inverted for both, which explains why when both are hybridized together, the male offspring are sterile
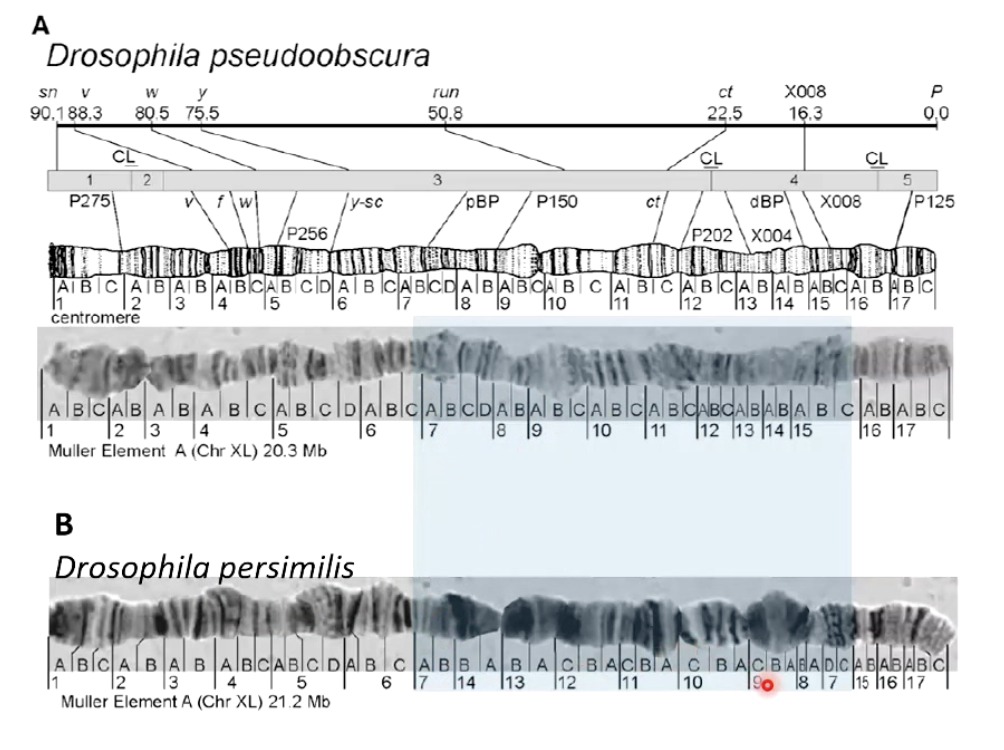
What is nondisjunction?
Failure of chromosomes or sister chromatids to separate properly during cell division.
Leads to chromosome number abnormalities (aneuploidy).
Can occur in Meiosis I or Meiosis II, producing different outcomes.
Fusion of gametes with normal gamete can produce trisomic (2n+1) or monosomic (2n-1) offspring
What is aneuploidy, and how does it occur?
Condition where an individual has an abnormal number of chromosomes.
Caused by nondisjunction during meiosis.
Example: Down syndrome (trisomy 21).
What happens when the X chromosome fails to separate?
Normal segregation: Each gamete gets one X.
Nondisjunction in Meiosis I: Gametes get two Xs or none.
Nondisjunction in Meiosis II: Gametes get two, one, or no Xs.
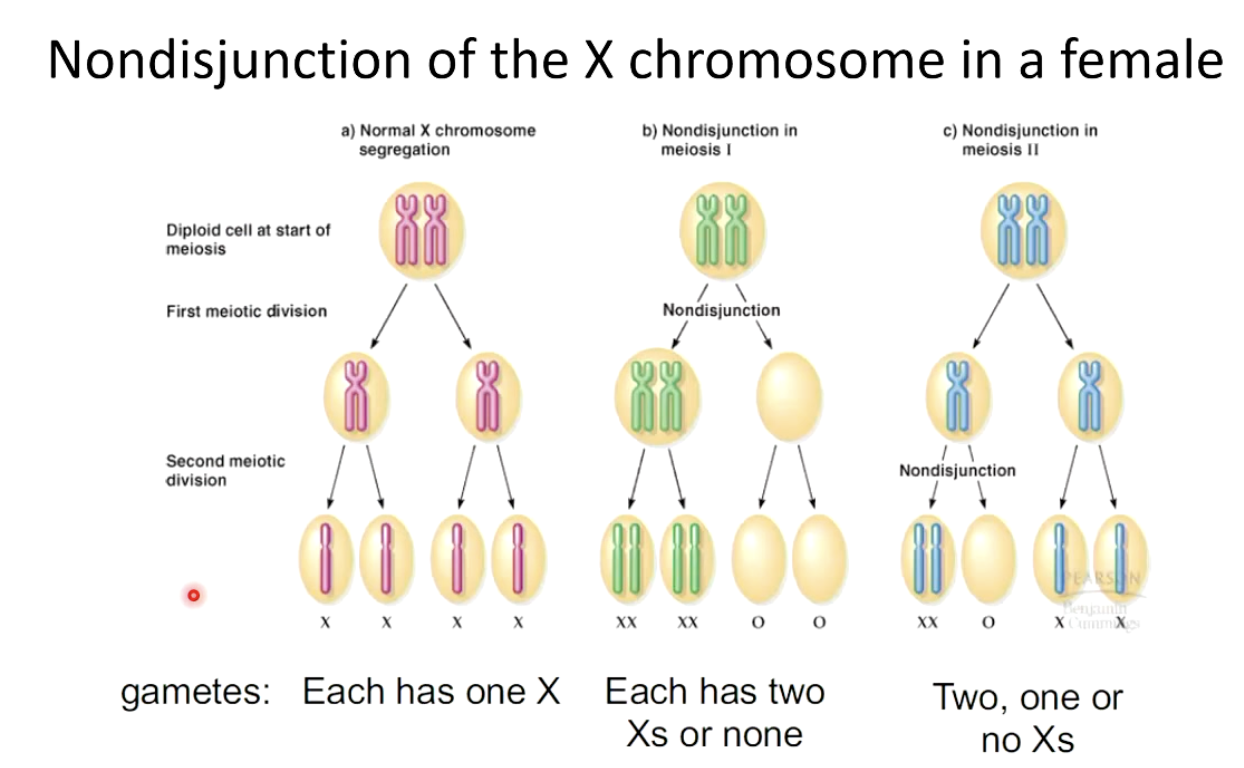
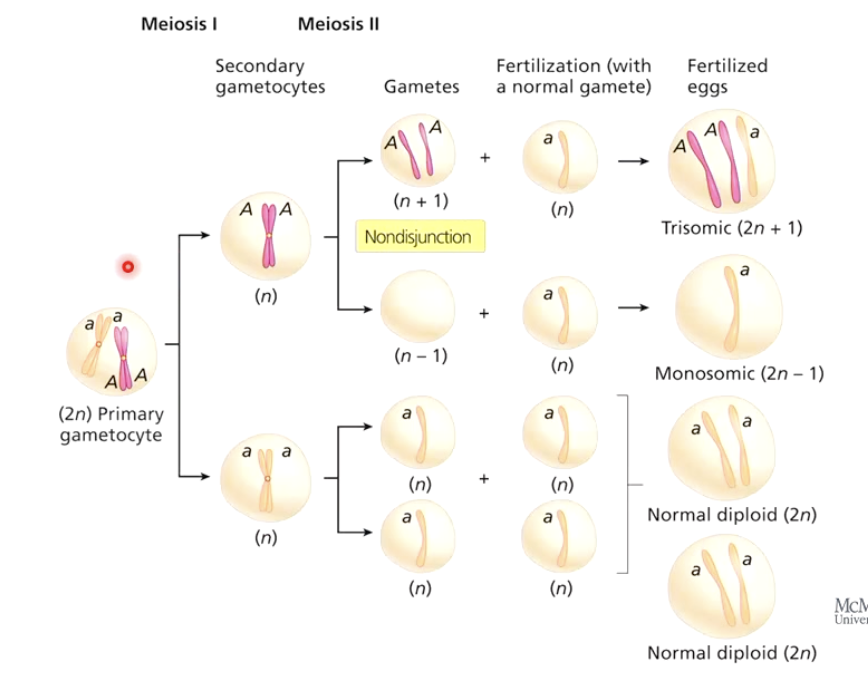
What happens during nondisjunction in Meiosis I?
Homologous chromosomes fail to separate.
One daughter cell gets an extra homolog, the other lacks a homolog.
Results: Four abnormal gametes.
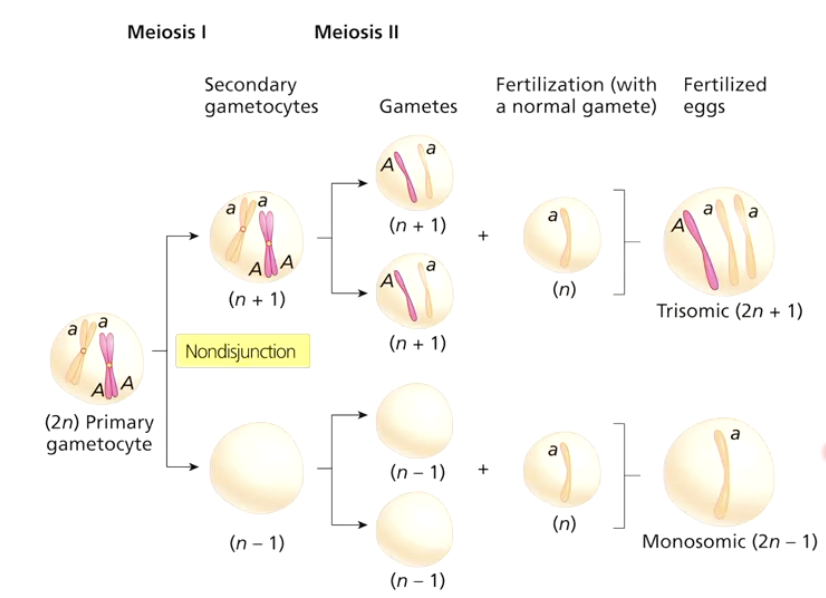
What happens during nondisjunction in Meiosis II?
Homologs separate normally, but sister chromatids fail to separate.
One daughter cell lacks a homolog.
Results: Two normal and two abnormal gametes.
How do humans and other animals respond to aneuploidy?
Highly sensitive to nondisjunction.
Most aneuploids do not survive gestation.
Certain syndromes result from aneuploidy.
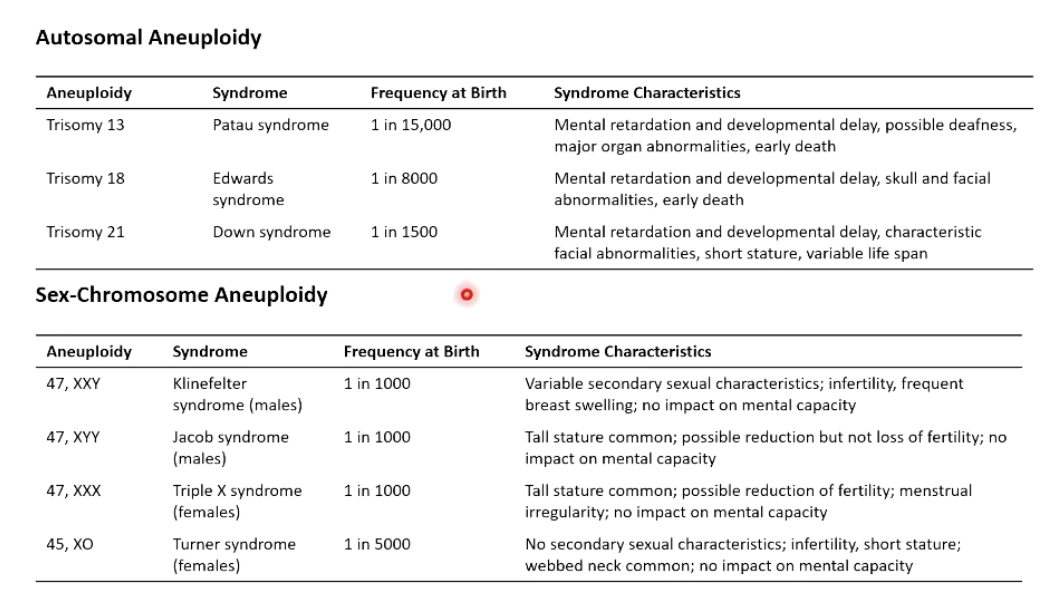
Autosomal trisomy can occur for chromosomes 13,18,21
No autosomal monosomies known - missing a chromosome
Sex chromosome trisonomies occur
X-chromosome monosomy can occur - Turner syndrome
Which autosomal trisomies are viable in humans?
Trisomy 13 (Patau syndrome)
Trisomy 18 (Edwards syndrome)
Trisomy 21 (Down syndrome)
No known autosomal monosomies.
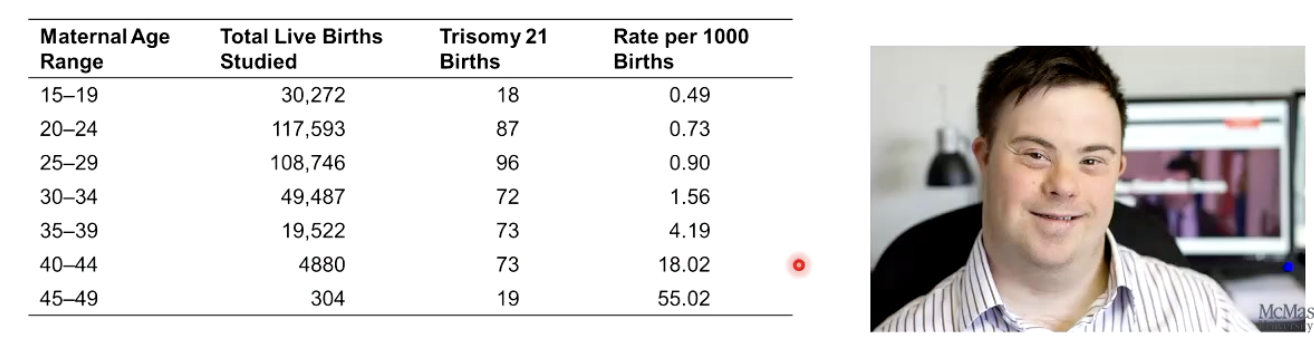
What is the most well-known and studied aneuploidy in humans?
Trisomy 21 (Down syndrome).
Most studied form of aneuploidy.
Increased risk with maternal age.
What is Robertsonian translocation?
Fusion of two non-homologous chromosomes into a large chromosome.
Can cause inherited Down syndrome.
Can affect segregation during Meiosis
Demonstrates Down syndrome is not always a result of a random non-disjunction.
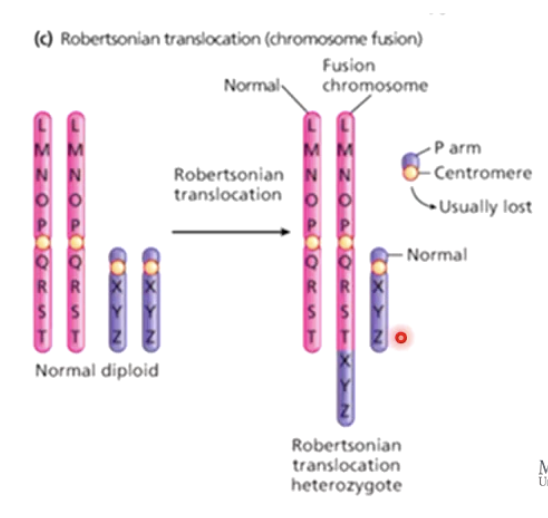
How can Robertsonian translocation lead to Down syndrome?
Fusion between chromosome 14 and 21.
Carrier has no symptoms but higher miscarriage risk.
If a fused chromosome + normal chromosome 21 are inherited, trisomy 21 results.
What is polyploidy?
Presence of three or more sets of chromosomes.
Common in plants.
What are the different levels of polyploidy?
Triploid (3n)
Tetraploid (4n)
Pentaploid (5n)
Hexaploid (6n)
Octaploid (8n)
What is the difference between autopolyploidy and allopolyploidy?
Autopolyploidy: Duplication of chromosomes within a species.
Allopolyploidy: Combining chromosome sets from different species.
What causes autopolyploidy and allopolyploidy?
Meiotic nondisjunction: Produces diploid instead of haploid gametes.
Example: 2n (egg) + n (pollen) = 3n plant.
Example: 2n (egg) + 2n (pollen) = 4n plant.
Mitotic nondisjunction: Doubles chromosome number after fertilization.
Both mechanisms can combine:
Example: 2n (egg) + n (pollen) = 3n plant → 6n plant after mitotic nondisjunction
How does autopolyploidy affect strawberries?
Small strawberry: Diploid (2n = 14).
Large strawberry: Octaploid (8n = 56).
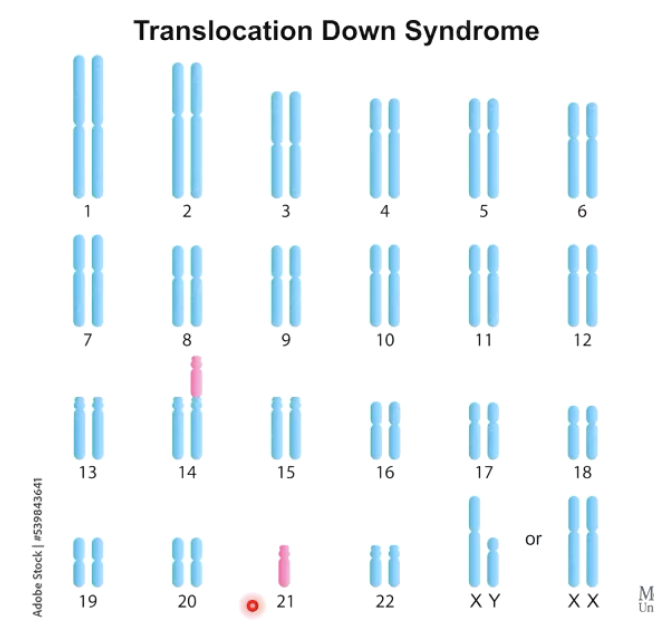
How does allopolyploidy create new species?
Chromosome number differs slightly between two species.
Fusion of gametes leads to 2n=61 chromosomes (odd number)
Mitotic nondisjunction doubles chromosome count → creates even number (e.g., 2n = 122).
Results in reproductively isolated species (2n=122) that are now fertile and can reproduce → creates new plant species.
How does chromosome breakage cause mutations?
Loss, gain, or rearrangement of chromosome regions.
Affects multiple genes, often with negative consequences.
Some changes have no effect (e.g., Robertsonian translocation).
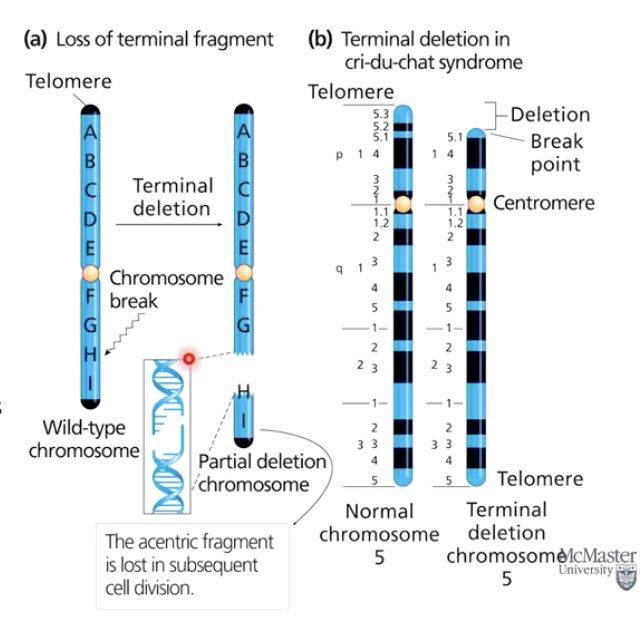
What happens when a chromosome breaks?
Both DNA strands break at a chromosome break point.
A terminal deletion occurs if an entire arm or part of it is lost.
Cri-du-chat syndrome results from a terminal deletion.
Acentric fragments (no centromere) are usually lost.
Interstitial deletions affect internal chromosome regions.
What happens when broken chromosome ends reattach incorrectly?
Inversion: Wrong ends reattach within the same chromosome.
Translocation: Reattachment occurs to a non-homologous chromosome.
If critical genes or regulatory regions remain unaffected, no phenotypic change may occur.
What are the two types of chromosome inversions?
Paracentric inversion: Centromere outside the inversion.
Pericentric inversion: Centromere within the inversion.
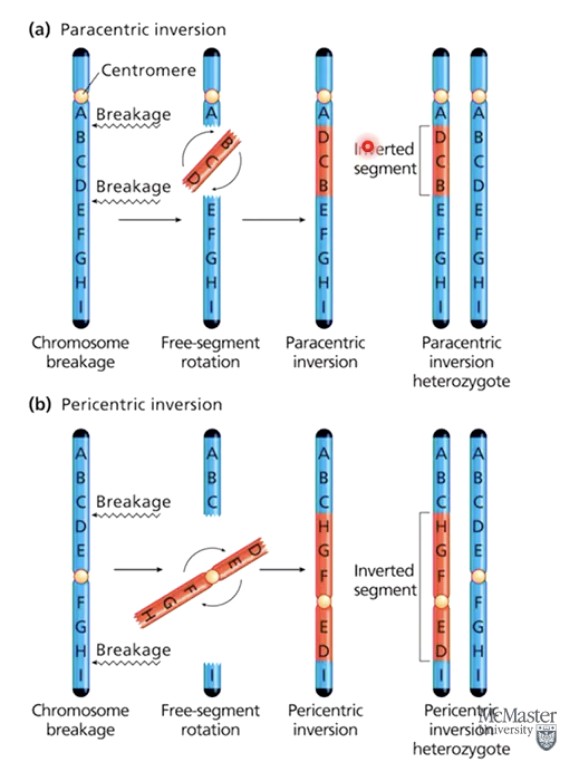
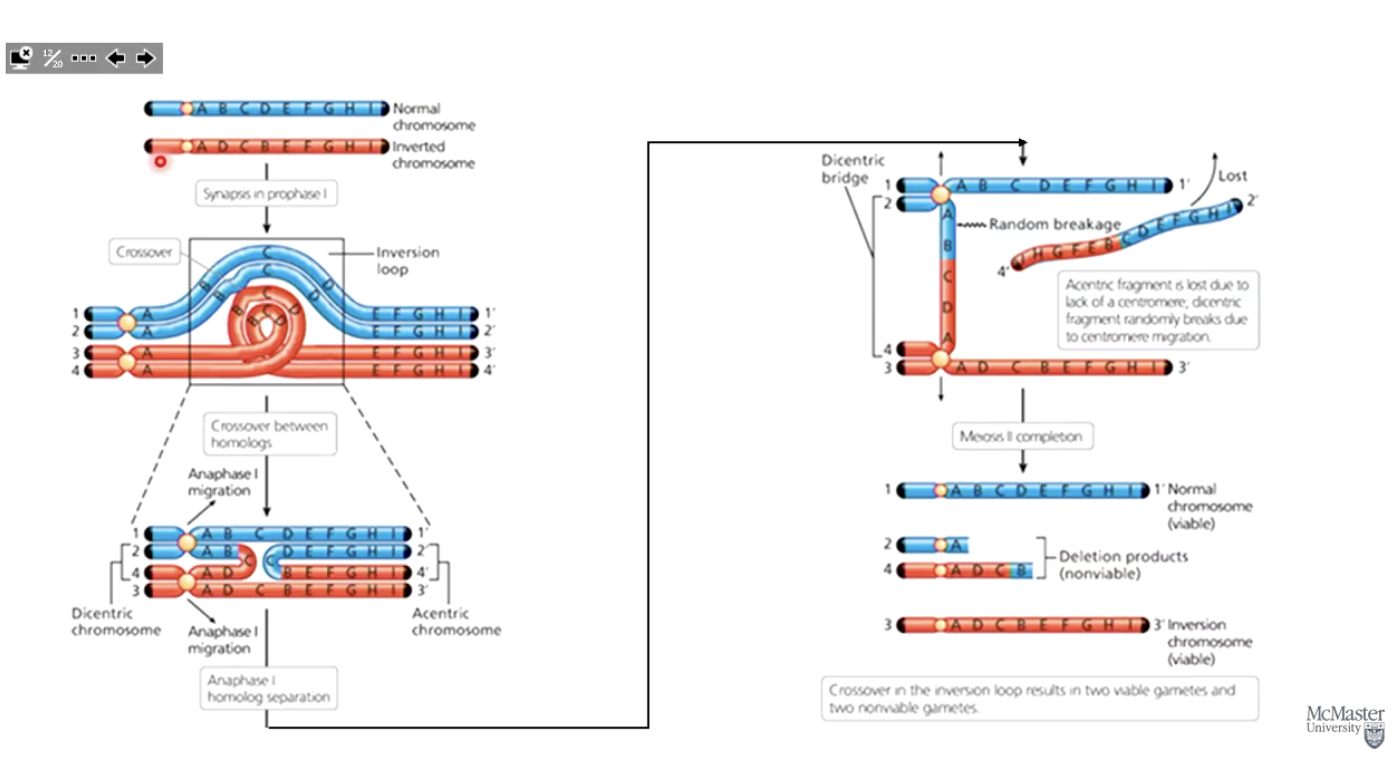
How do inversions affect recombination?
Suppress recombination in heterozygotes.
Requires homologous regions to exchange DNA.
Inversion heterozygotes have one normal and one inverted chromosome, disrupting homology.
What happens when recombination occurs in an inversion heterozygote?
A physical inversion loop forms between chromosomes.
Crossover in the inversion loop can cause chromosome breakage.
Resulting gametes have large deletions, usually leading to inviable offspring when fertilized with a normal gamete.
Photo: Blue is normal chromosome, red is inversion.
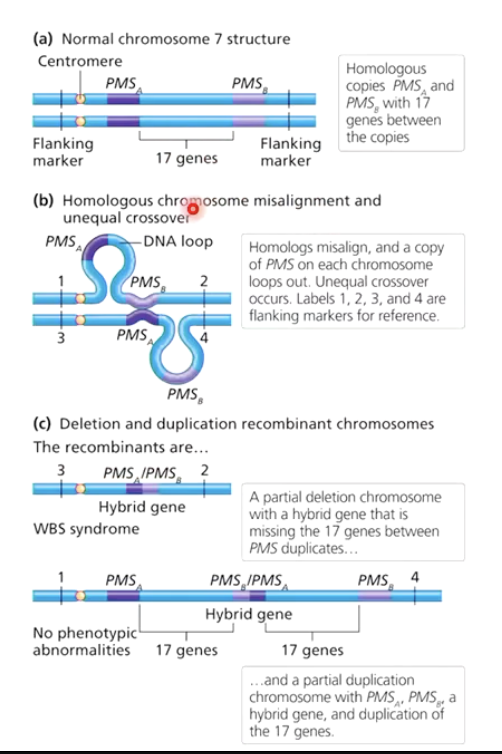
What is unequal crossover?
Rare crossover event between homologs.
Results in partial duplication on one homolog and partial deletion on the other.
Example: Williams-Beuren syndrome from unequal crossover on chromosome 7.
What genetic event causes Williams-Beuren syndrome?
Duplication of the PMS gene on chromosome 7 due to unequal crossover.
Symptoms: Naïve, overly trusting personality, mild intellectual disabilities, heart problems.
What is deletion mapping?
Genetic technique used to map genes by comparing phenotypes in mutants with known deletions.
Often used in model organisms like Drosophila with well-mapped deficiency lines.
Widespread in flies because they have dosage compensation mechanisms on their X-chromosomes and autosomes so they can tolerate having deletions on one copy
What are the key steps in deletion mapping?
Note: Requires mutation to be recessive
Cross mutant of interest with strains carrying known deletions.
Look for pseudodominance (if organism has a recessive allele on one chromosome, and deletion on the other, the recessive allele is expressed).
Compare to genetic markers to pinpoint gene location.
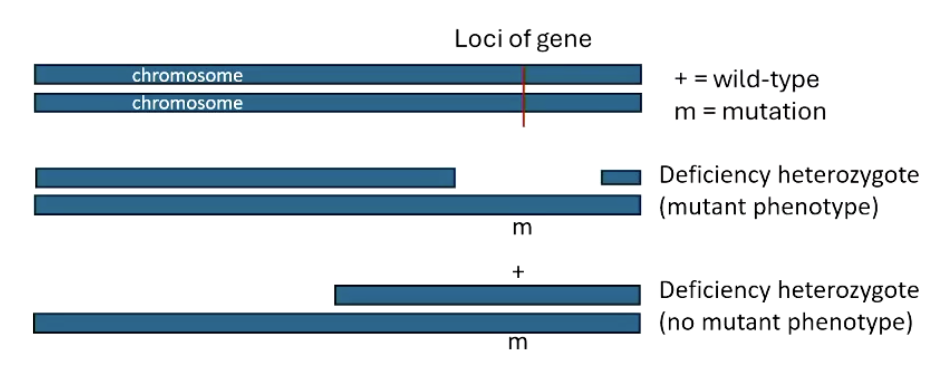
What is pseudodominance, and why is it important for deletion mapping?
Pseudodominance: Expression of a recessive allele when the corresponding dominant allele is deleted.
Helps identify gene location by mimicking hemizygous conditions.
What did deletion mapping reveal about the Drosophila notch gene?
notch is a developmental gene affecting wing phenotypes.
Deletion mapping narrowed it down to a specific region on the X chromosome.
Photo: Blue shows where the deletion is, recessive notch mutation (pink) is expressed where it overlaps with the blue deletion
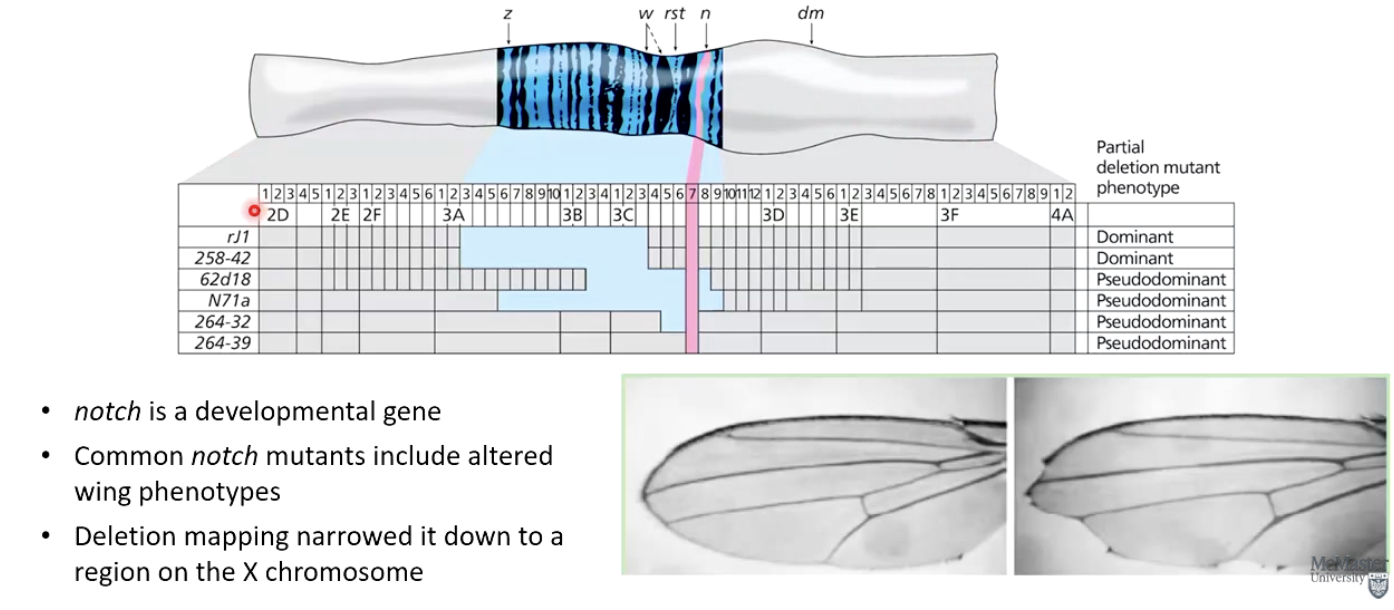
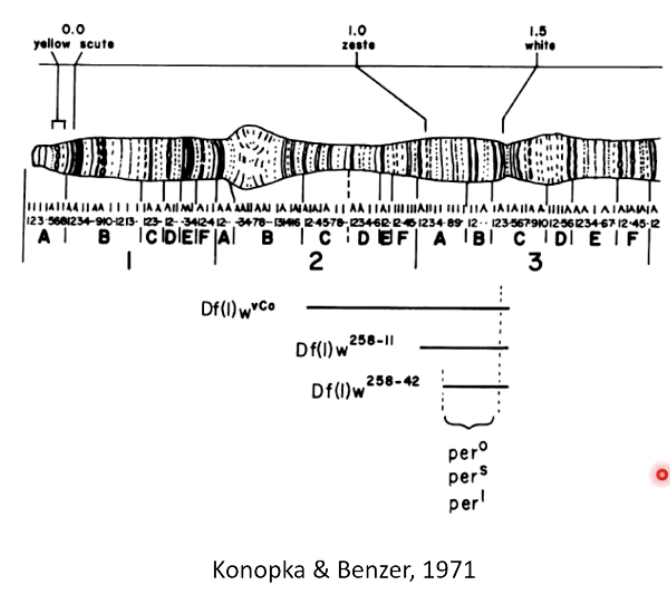
How did Konopka & Benzer (1971) use deletion mapping?
Studied mutants with altered circadian rhythms.
Used deletion mapping to identify the first circadian clock gene on the X chromosome.
Used a genetic map with well-known genetic markers (e.g., yellow and white genes).