MICR5831 L14: Motility and Chemotaxis 10/1/25
1/87
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
88 Terms
What is this?
-Nanomachine a millionth the size of a grain of sand
-Rotates five times faster than a Formula One engine
-Changes direction faster than a mosquito beats it wings
Flagella
What are some examples where motility is associated with virulence?
-H. pylori
-V. cholerae
-Salmonella and Campylobacter
What is this?
-Helical thread-like appendages
-Extend from the plasma membrane and cell wall
-Rotation acts like the propeller of a boat
-Slender rigid structures
Flagella
True or False: Flagella are straight
False
True or False: Flagella have a constant wavelength associated with their species
True
How can you prove flagella filament rotation acts like the propeller of a boat?
If the flagellum is artificially tethered at the tip, bacterial cell rotates
What is this?
-Flagella "on both sides"
-Single flagella at opposite ends of cell
Amphitrichous
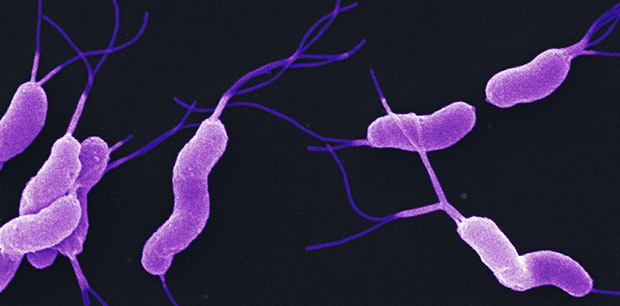
What is this?
-Flagella "tuft"
-Cluster at one or both ends
-Associated with H. pylori
Iophotrichous
What is this?
-"Around"
-Flagella spread over cell-surface
Petririchous
What are the 3 parts of a Flagellar structure?
1) Filament
2) Hook
3) Basal body
Which part of the flagella is this?
-Hollow, rigid cylinder
-Composed of ~20,000 flagellin protein molecules
-Synthesized by transfer of flagellin subunits through the hollow filament (spontaneous aggregation occurs)
Filament
Which part of the flagella is this?
-Over 40 genes (fla, fli and flg)
-Control synthesis and motility
-Synthesis involves 20-30 genes
Filament
What is this?
-Filament
-Hollow, rigid cylinder
-Composed of ~20,000 flagellin protein molecules
-Synthesized by transfer of flagellin subunits through the hollow filament (spontaneous aggregation occurs)
Which part of the flagella is this?
-Composed of protein
-Acts as a flexible coupling between the filament and basal body
Hook
What is this?
-Hook
-Composed of protein
-Acts as a flexible coupling between the filament and basal body
Which part of the flagella is this?
-Four rings attached to a central rod
-Attaches flagellum to the cell
-Functions as the flagellum motor
Basal body
What is this?
-Basal body
-Four rings attached to a central rod
-Attaches flagellum to the cell
-Functions as the flagellum motor
What are the Basal body's 4 rings?
-L, MS, P, C
-Lipopolysaccharide
-MS (Plasma membrane)
-Peptidoglycan
-Cytoplasmic side ring
How are C and MS motor rings attached?
Cytoplasmic sides
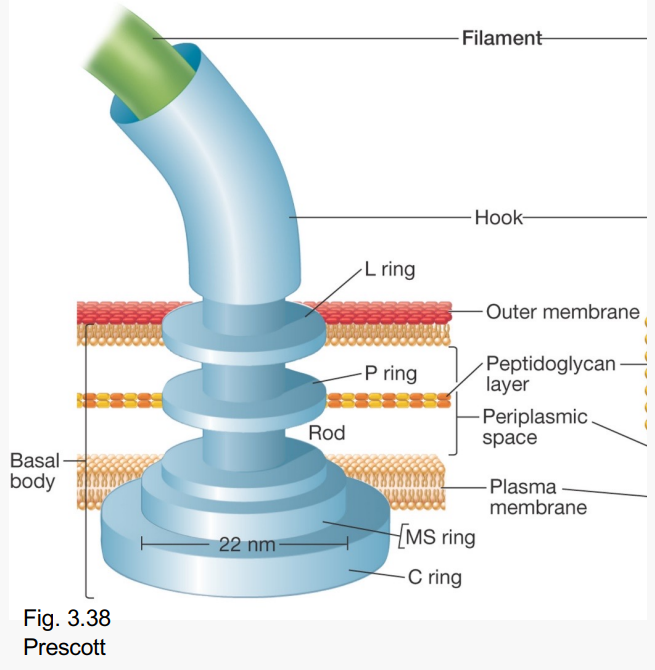
What do these Basal body rings do?
-P and L
Act like bearings
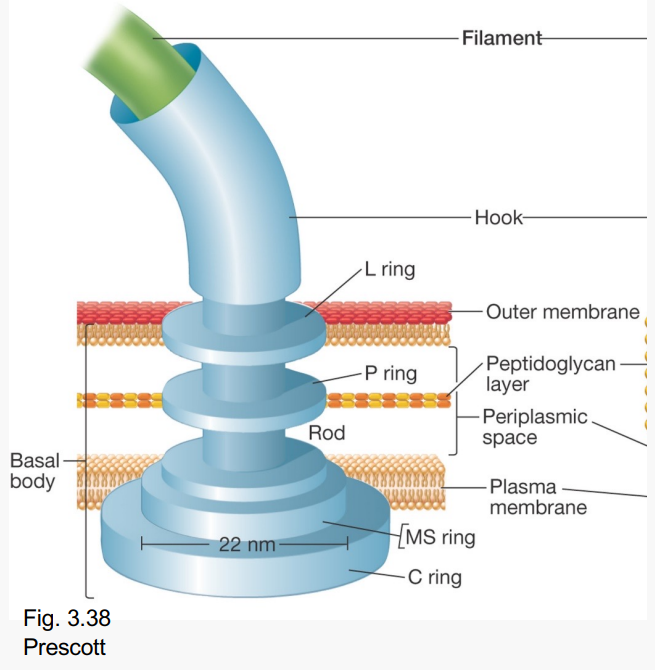
What do these Basal body rings do?
-MS and C
-Acts as motor
-Rotates within membrane
-Turns attached rod/filament
Basal bodies are found in Gram ____ bacteria
Negative
What is this?
-Drives flagellar rotation
-H+ flow occurs at Mot protein channel
-Protons move down a charge and pH gradient
Proton Motive Force (PMF)
What gives PMF the push?
-Proton enters Mot channel
-1000 protons required for complete rotation
Why do protons entering the Mot channel cause it to rotate?
Protons exert electrostatic forces on charges present in C and MS rings
True or False: Rotation speed is proportional to the PMF
True
What happens if flagellar rotation happens counterclockwise?
Run, straight swim
What happens if flagellar rotation happens clockwise?
Tumble, jiggle, random change of direction
What is this?
-Cells swim forward or backward by reversing flagellar rotation
-CCW rotation pushes, CW rotation pulls the cell
Polar flagella
What is this?
-Counterclockwise rotation causes running
-Clockwise rotation causes tumbling (disrupts a run)
-Cell moves off in a new direction
Petririchous flagella
What motion does this describe?
-Forward swim is linear
-Backward swim is in a curve
Run and arc
What motion does this describe?
-Sudden bending of hook and filament
-"Flick" of the flagellum after the reverse swim
Run-reverse-flick
What motile bacteria species is this?
-Rotates 1100 rps
-Speeds of up to 90 µm per sec or 60 cell lengths/sec
-Equates to a human running speed of ~ 40 km/hr
Vibrio alginolyticus
True or False: In a homogeneous environment, the direction of bacterial motility is random
True
What flagellar motility would you expect from this?
-No nutrient gradient
-Straight runs are interspersed with tumbles
-Result in random directional changes when running resumes
What are some taxis responses?
-Chemotaxis (chemicals)
-Aerotaxis (oxygen)
-Phototaxis (light)
-Thermotaxis (temperature)
-Osmotaxis (water potential)
-Magnetotaxis (magnetic fields)
What is this?
-Bacteria can respond to remarkably low levels of chemicals (e.g. 10 nM for some sugars)
-They detect concentration gradients as small as a change of 1 molecule per cell volume per µm, while being buffeted by Brownian motion in liquids
Chemotaxis
What flagellar motility would you expect from this?
-Nutrient gradient
-Biased random walk
-Fewer tumbles when running up gradient towards nutrients
-Tumbling occurs at normal intervals travelling down gradient
How do prokaryotic cells sense concentration changes?
Periodic/temporal sampling of the environment
What is this?
-Chemoreceptors that detect attracts and repellents
-Located in the plasma membrane
-Integral or transmembrane proteins
Methyl-accepting chemotaxis proteins (MCPs)
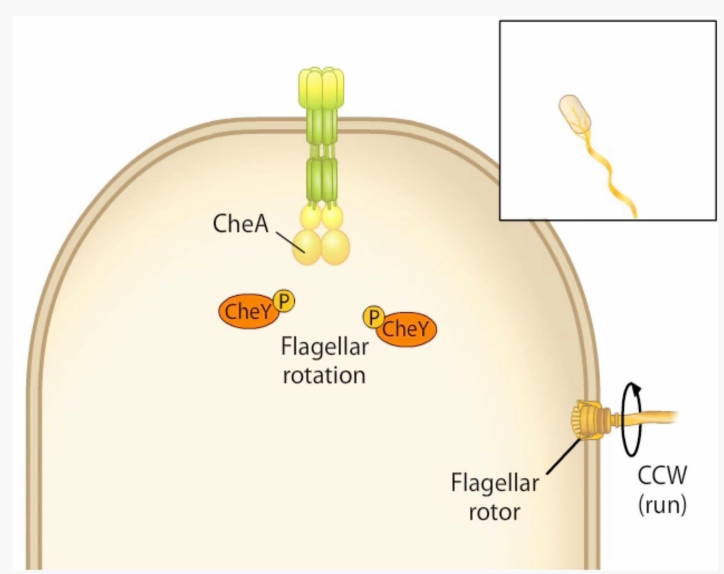
What key protein for signal recognition/chemotaxis is this?
-Histidine Kinase (HK)
-Associates with a number of MCPs via CheW adaptor protein
-Phosphorylates CheB methylesterase and CheY
-Switches from CCW → CW rotation
CheA
True or False: More methylation = more attractant required for CheA kinase activity
True
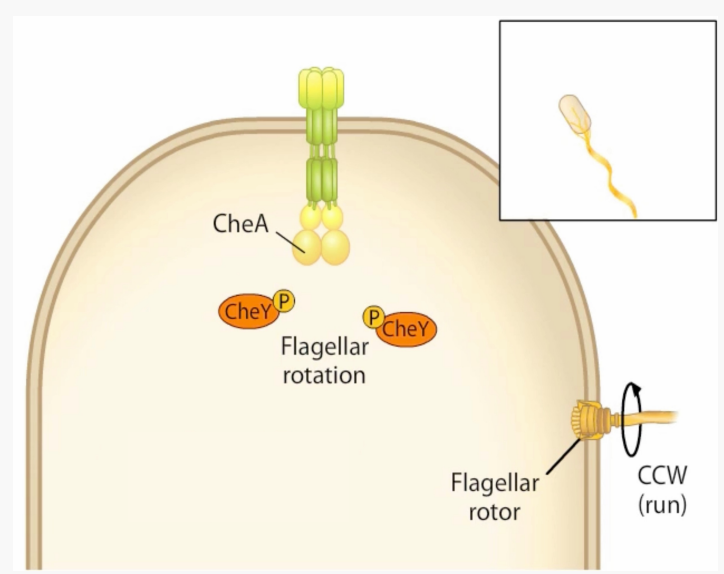
What key protein for signal recognition/chemotaxis is this?
-Methylesterase that removes methyl groups
-Resensitises MCP to lower levels of attractant
-Activated by phosphorylation by CheA
CheB
What key protein for signal recognition/chemotaxis is this?
-Removes methyl groups from MCPs
-Phosphorylated by CheA in absence of attractants
-Resensitizes MCPs to lower attractant levels
CheB-P
True or False: Methylation and demethylation of MCPs occurs slower than the phosphorylation
True
Which two MCPs are phosphorylated by CheA?
1) CheB
2) CheY
What happens after demethylation and resensitization to lower attractant levels?
-Cell returns to normal swim/tumble frequency
-Can respond to increasing attractant levels again
What key protein for signal recognition/chemotaxis is this?
-MethyltransfeRase
-Adds methyl groups (unlike methylesterase which removes)
-Adapts MCPs to higher amounts of attractant
-Each MCP can be methylated by up to 4 methyl groups
CheR
What key protein for signal recognition/chemotaxis is this?
-Adaptor protein
-Helps CheA to couple/associate with MCPs for phosphorylation
CheW
What key protein for signal recognition/chemotaxis is this?
-Phosphorylated by CheA when it is not bound to attractant
-Activated form associates with flagella and CW = tumble
CheY
True or False: Methyl-accepting chemotaxis proteins (MCPs) bound by attractant are activated
False
True or False: MCPs bound by attractant are inactivated, suppressing tumbles (less random motion)
True
What key protein for signal recognition/chemotaxis is this?
-Active, phosphorylated protein
-Directly related to rate of tumbles
-Binds to the flagellar motor
-Switches rotation to CW = tumble
CheY-P
What key protein for signal recognition/chemotaxis is this?
-Phosphatase
-Dephosphorylates CheY in absence of attractants
-Results in mainly CCW = run
CheZ
What is the default flagellar rotation?
-CCW = swim
-Chemotaxis signal transduction regulates tumbles (switch to CW rotation)
How does signal transduction drive CW flagellar rotation (tumbles)?
1) CheA phosphorylates CheY when MCPs do not sense any attractant
2) CheY-P binds the flagellar motor and switches direction to CW (tumbles)
3) MCP encounters attractant, then eventually runs out of it
4) CheZ dephosphorylates CheY-P in absence of attractant and inactivates it resulting in CCW = run
Name the first step of signal transduction driving CW flagellar rotation (tumbles)
1) CheA phosphorylates CheY when MCPs do not sense any attractant
Name the 2nd step of signal transduction driving CW flagellar rotation (tumbles):
1) CheA phosphorylates CheY when MCPs do not sense any attractant
CheY-P binds the flagellar motor and switches direction to CW (tumbles)
Name the 3rd step of signal transduction driving CW flagellar rotation (tumbles):
2) CheA phosphorylates CheY when MCPs are unbound by attractant
MCP switches to CW tumbling and encounters attractant, eventually runs out of it
Name the 4th step of signal transduction driving CW flagellar rotation (tumbles):
3) MCP switches to CW tumbling, encounters attractant, then eventually runs out of it
CheZ dephosphorylates CheY-P in absence of attractant and inactivates it, resulting in CCW = run again
How does MCP methylation adapt bacteria to higher chemoattractant concentrations?
1) CheR methylates MCPs to require higher concentrations of attractant, prefers bound MCPs
2) CheB-P removes methyl groups in lower concentration of attractant to resensitize MCPs
Which does CheR prefer for methylating, unbound or bound MCPs?
Bound
Which motile bacterial flagella is this?
-lophotrichous cells have tuft of 4-7 flagella
-Causes inflammation of the stomach lining
-Linked to gastric ulcers and stomach cancer
Helicobacter pylori
Which motile bacterial flagella is this?
-Flagellum-negative fla- mutants
-Unable to colonise gnotobiotic (germ-free) piglets' stomachs
Helicobacter pylori
Which motile bacterial flagella is this?
-Cells comma-shaped, have single polar flagellum
-Infects small intestine w/ profuse watery diarrhea, vomiting, rapid dehydration, electrolyte loss
Vibrio cholerae
Which motile bacterial flagella is this?
-Flagellum-negative (Fla-) mutants had reduced virulence in animal models but were still capable of colonization
Vibrio cholerae
What chemotaxis is this?
-Spiral cells w/ very rapid "corkscrew" motion
-In high viscosity fluids e.g. mucus
-Products will inflame and damage the mucosa
Helicobacter pylori
What chemotaxis is this?
-Urea is the main attractant, attracted to epithelial layer
-Swim down through chemotaxis and colonize the mucin layer
-Adhere to gastric mucosa, colonizing gastric pits
Helicobacter pylori
What is this?
-Used by H. pylori as protection from stomach in mucin layer
-Increases pH, converts substrate to Ammonia + Bicarbonate
Urease
What chemotaxis is this?
-Flagella swim through mucous layer to the intestinal wall
-Fla+ CW mutant with decreased virulence
-Fla+ CCW with increased virulence
Vibrio cholerae
What happens if a Fla+ Vibrio cholerae mutant has the following mutation?
-CW: Flagella rotated clockwise only (tumble only)
Mutant had decreased virulence
What happens if a Fla+ V. cholerae mutant has the following mutation?
-CCW: Flagella rotated counterclockwise only (straight run only)
Mutant had increased virulence
Describe the CCW only pathway of a Vibrio cholerae mutant
1) Straight runs
2) Able to cross lumen of small intestine (swim random directions, but some towards villi)
3) Some bacteria rapidly make contact with villi of SI, bind and colonize host
4) Increased virulence
Describe the CW only pathway of a Vibrio cholerae mutant
1) Tumbles
2) Unable to get to receptors on intestinal villi
3) Decreased virulence
Why would Vibrio cholerae not evolve to always exhibit its max possible colonization?
-Vibrio cholerae in rice-water-stools exists in transient state of nonchemotaxis
-Vibrio cholerae accumulates in the lumen
-Increases its chances of infecting new hosts
-No need to evolve further
What is the 1st stage of V. cholerae infection?
1) Bacteria enter GIT
-Bile in the lumen keeps most virulence genes expression OFF, but enhances motility
-2-component transcriptional regulatory system
What is the 2nd stage of V. cholerae infection?
1) Bacteria enter GIT
2) Bacteria swim into the mucous gel
-Bile concentration is reduced
-Virulence genes ON, motility decreased
-Increase in viscosity also sensed by bacteria
-Reduces flagella production (mechanosensing)
What is the 3rd stage of V. cholerae infection?
2) Bacteria swim into the mucous gel
3) Bacteria colonize epithelium
-Bacteria produce Toxin Co-regulated Pilus (TCP) adhesins
-Bind to epithelial cell brush border
-Secrete Cholera Toxin (CT)
-Nonmotile mutants pass through without colonization
Which stage of V. cholerae infection is this?
-Bile in the lumen enhances motility
-Bile keeps most virulence gene expression OFF
-2 component transcriptional regulatory system
1) Bacteria enter GIT
Which stage of V. cholerae infection is this?
-Bile concentration is reduced
-Virulence genes ON, motility decreased
-Increase in viscosity also sensed by bacteria
-Reduces flagella production (mechanosensing)
2) Bacteria swim into mucous gel
Which stage of V. cholerae infection is this?
-Bacteria produce Toxin Co-regulated Pilus (TCP) adhesins, bind to epithelial cell brush border and secrete Cholera Toxin (CT)
3) Bacteria reach brush border and colonize the epithelium
What do Salmonella and Campylobacter have in common?
1) Flagella utilization during infection is independent of motility
2) Both cause diarrhea and SI inflammation
What is this?
-Fla- mutants have reduced virulence in an animal model
-Non-motile Fla+ mutants have normal virulence
-Flagella are independent of motility
-Flagella are not heavily glycosylated
Salmonella
What is the role of flagella in Salmonella?
-Flagellum acts as an Adhesin
-Important in biofilm formation
-Independent of motility
What is this?
-Glycans mediate bacterial autoagglutination and microcolony formation
-Promoting adhesion to intestinal cells and biofilm formation
Campylobacter
What sets Campylobacter flagella apart from Salmonella?
-Glycans
-Flagellin is heavily glycosylated
Why would Campylobacter use glycans to mediate adhesion and biofilm formation?
-Mask the flagellum from the immune response
-Flagella are highly immunogenic
What is the role of flagella in Campylobacter?
-Autoagglutination, microcolony formation
-Adhesion and biofilm formation
-Independent of motility