BIO 311 Exam #1
1/123
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
124 Terms
allosteric regulation
bind to the enzyme → conformational change → decrease enzyme activity
feedback inhibition: regulate levels of the synthesized end product
reversible
covalent modification
add phosphate groups → add negative charge and change electrostatic properties
reversible
proteolytic modification
digestive enzymes, clotting factors
synthesized as inactive pro-enzymes, cut up, segments activated
not reversible
catabolism
degradation
release energy
creates ATP, NADH, NADPH, FADH2
exergonic processes
anabolism
synthesis
uses energy
ATP, NADH, NADPH, FADH2
endergonic processes
heterotroph
use C from food
energy from degradation
makes CO2, H2O
autotroph
use C from air (CO2)
energy from sunlight
make O2, H2O
oxidation
loss of electrons
losing electrons = reducing agent
catabolism
reduction
gaining electrons
gaining electrons = oxidizing agent
anabolism
FAD/FADH2 and FMN/FMNH2
act as coenzymes in many enzyme-catalyzed RedOx reactions
can accept 1 or 2 hydrogens
redox common in catabolism
substrate undergoes oxidation
lose 2H (2 p+ and 2 e-)
oxidized form of NAD accepts hydride
:H- (1 p+, 2e-)
redox common in anabolism
NADH donates hydride to oxidized substrate
substrate becomes reduced
lactate → pyruvate
lactate dehydrogenase
2e- and 2H+ removed from C2 of lactate (alcohol) to make pyruvate (ketone)
reversible
homolytic cleavage
each atom keeps one of the bonding electrons
heterolytic cleavage
one of the atoms keeps both of the bonding electrons
isomerization
redistribution of electrons within a molecule
can result in isomerization, transposition of double bonds, cis-trans rearrangement
isomerases
eliminations
elimination of water
introduce C=C between 2 saturated Cs
acyl group transfer
addition of nucleophile to carbonyl C to form a tetrahedral intermediate
phosphoryl group transfer
attachment of goof LVG to metabolic intermediate to “activate” the intermediate for subsequent reactions
not simple ATP hydrolysis
formation of phospho-substrate intermediate gives greater free energy
displacement of P group from substrate
glycolysis
cytoplasm
oxidation of glucose → pyruvate
creates ATP, NADH
TCA cycle
mitochondria
pyruvate → citrate
creates NADH, FADH2, CO2
electron transfer & oxidative phosphorylation
mitochondria
p+ pumps and e- transfer drive ATP synthesis
creates ATP, H2O
sucrose
1,2-linked α-glucose + β-fructose
α-1,2-glycosidic linkage
source: sugar cane
lactose
1,4-linked β-galactose + α-glucose
β-1,4-glycosidic linkage
source: milk
maltose
1,4-linked α-glucose + α-glucose
α-1,4-linked glycosidic bond
source: hydrolyzed starch
why can’t we digest cellulose?
humans do not have the enzymes that can break β-1,4-glycosidic bonds
cellulose is too large for lactase to accommodate in its active site
amylopectin
more branching, α-1,4-linked glucose with α-1,6-linkages at branch points
amylose
long, unbranched chains of D-glucose; α-1,4 linked
glycogenin reactions
attachment of UDP-glucose to glycogenin protein
transfer of glucosyl residues to existing (glu)n-glycogenin
glycogen breakdown steps
(glycogen)n glu + Pi → (glycogen)n-1 glu + G1P
G1P → G6P
G6P + H2O → glucose + Pi
removal of branch points
glycogen breakdown - enzymes
glycogen phosphorylase
phosphoglucomutase
glucose-6-phosphatase
debranching enzyme
reducing sugars
sugars that reduce mild oxidizing agents
debranching enzyme
remove all but one glucosyl residue from a branch point and transplants the short chain to neighboring branch
α-1,6-glucosidase: removes last glucosyl residue as glucose
glycogen synthase
(glycogen)n glu + UDPG → (glycogen)n+1 glu + UDP
glycogen synthesis
glucose + ATP → G6P + ADP
G6P → G1P
UTP + G1P → UDP-glucose + pyrophospate
UDP-glu + (glycogen)n glu → (glycogen)n+1 glu + UDP
pyrophosphate + H2O → 2Pi + H+
branching enzyme
glycogen synthesis enzymes
hexokinase (glucokinase in the liver)
phosphoglucomutase
UDP-glucose pyrophosphorylase
pyrophosphatase
branching enzyme
branching enzyme
glycogen synthase makes α-1,4 glycosidic bonds
branching enzyme transplants short chain to introduce branch by forming an α-1,6-glycosidic bond
insulin
lowers blood glucose
inhibits glycogen breakdown (promotes synthesis)
inhibits gluconeogenesis
epinephrine and glucagon
raise blood glucose
stimulate glycogen breakdown
stimulate gluconeogenesis
low blood glucose: increased glycogen breakdown
low blood glu
inc glucagon
inc cAMP
inc PKA
PKA phos + activates phosphorylase kinase
inc phosphorylase kinase phos + activates glycogen phosphorylase
inc glycogen phosphorylase
low blood glucose: decreased glycogen synthesis
low blood glu
inc glucagon
inc cAMP
inc PKA phos + deactivates glycogen synthase
dec glycogen synthase
high blood glucose: decreased glycogen breakdown
inc insulin
inc insulin-sensitive protein kinase
inc PP1
PP1 dephos + inactivates phosphorylase kinase
dec phosphorylase kinase leads to dec glycogen phosphorylase (inactive)
high blood glucose: increased glycogen synthesis
inc insulin
inc PKB
PKB phos + inactivates GSK3
inactive GSK3 leads to dephosphorylated glycogen synthase (active)
more glycogen synthase
allosteric effects: ATP
G6P
high E state
inhibit phosphorylase
allosteric effects: AMP
activate phosphorylase
E state of cell is low
glycogen breakdown
insulin & GM protein
phosphorylation of GM site 1
activate PP1
dephosphorylates glycogen phosphorylase kinase, glycogen phosphorylase, and glycogen synthase
epinephrine & GM protein
phos of Gm site 2
PP1 dissociates from glycogen
prevent PP1 access to glycogen phosphorylase & glycogen synthase
products of glycolysis
per glucose molecule:
2 ATP
2 NADH
2 pyruvate
3 possible fates for pyruvate
aerobic respiration (CO2 + H2O)
anaerobic respiration (lactate)
anaerobic respiration (ethanol)
glycolysis reaction overall
glucose + 2NAD+ + 2ADP + 2Pi → 2 pyruvate + 2NADH + 2H+ + 2ATP + 2H2O
glycolysis prep phase
4 steps
converts 6C sugar to 2 3C sugars
uses 2 ATP
glycolysis payoff phase
6 steps
converts 2 3C sugars to 2 pyruvate
makes 4 ATP (2 from each 3C sugar)
glycolysis step 1
glucose → G6P
phosphorylation
ATP → ADP
hexokinase
priming reaction
rate limiting step
“traps” glucose as G6P which does not diffuse out of cells or bind to glucose transporters
glycolysis step 2
G6P → F6P
phosphohexose isomerase
reversible
isomerization
glycolysis step 3
F6P → F-1,6-BP
PFK-1
ATP → ADP
phosphorylation
not reversible
first committed step
rate-limiting step
second priming step
glycolysis step 4
F-1,6-BP → DHAP + GAP
aldolase
reversible
cleavage
glycolysis step 5
DHAP → GAP
triose phosphate isomerase
reversible
glycolysis step 6
GAP → 1,3-BPG
GAP dehydrogenase
reversible
NAD+ → NADH + H+
generation of a high-energy compound
glycolysis step 7
1,3-BPG → 3-phosphoglycerate
phosphoglycerate kinase
2 ADP → 2 ATP
reversible
substrate-level phosphorylation
glycolysis step 8
3-PG → 2-PG
phosphoglycerate mutase
reversible
rearrangement
glycolysis step 9
2-PG → PEP
enolase
reversible
H2O released
generation of a high-energy compound
glycolysis step 10
PEP → pyruvate
pyruvate kinase
2 ADP → 2 ATP
not reversible
rate-limiting
substrate-level phosphorylation
fermantation
lactic acid, ethanol
energy extraction without consumption of oxygen
no net change in the [NAD+] or [NADH]
lactic acid fermentation
pyruvate → L-lactate
reversible
lactate dehydrogenase
NADH + H+ → NAD+
2 redox reactions; no net change in oxidation state of C in glucose
no net change in oxidation
2ATP/glucose extracted in conversion of glucose → lactate
ethanol fermentation
pyruvate → acetaldehyde
reversible
CO2 released
pyruvate decarboxylase
acetaldehyde → ethanol
reversible
NADH + H+ → NAD+
pasteur effect
yeast consume more sugar when grown under anaerobic conditions
hexokinase deficiency
reduced glucose breakdown
reduced ATP production
reduced BPG production
not as easy for Hb to assume T-state
pyruvate kinase deficiency
reduced ATP production
RBCs become deformed/lyse
Hb carries less O2
individuals with a deficiency in pyruvate kinase would be expected to display a decrease in hemoglobin affinity for oxygen
glycolysis & cancer
cancer cells grow more rapidly than blood vessels supplying them
hypoxic tumors express HIF-1
HIF-1 increases gene expression (glycolytic enzymes, GLUT)
HIF-1 stimulates growth of vasculature (VEGF)
anaplerotic reaction
reactions that replenish intermediates depleted by other reactions
generation of acetyl-CoA
CoA gets 2C from pyruvate in the form of an acetyl group
High-energy thioester linkage
Multi-enzyme process (coupling)
E1
pyruvate dehydrogenase
uses TPP as bound cofactor
attacks C2 of pyruvate, releases CO2
TPP remains bound to hydroxyethyl group
E2
dihydrolipoyl transacetylase
lipoic acid: cofactor covalently bound to Lys of E2
creates long, flexible arm - can move between active sites of all 3 PDH enzymes
disulfide reduced to SH + SH
lipoamide side chain extends to E1
transfers hydroxyethyl from TPP to dihydrolipoamide
partial reduction creates acetyl group
second reduction transfers acetyl group to CoA
E3
dihydrolipoyl dehydrogenase
resets the system
catalyzes the regeneration of disulfide (oxidized) form of lipoamide/lipolysine
uses bound cofactor (FAD)
NAD+ oxidizes FADH2 to regenerate FAD
NAD becomes reduced (NADH + H+)
PDH complex regenerated; NADH + H+ made
acetyl-CoA derived from FA
creating a fatty acyl-CoA
thiol group of CoA-SH carries out a nucleophilic attack
β-oxidation
each 4-step “pass” removes one acetyl group (2C) from chain to make acetyl-CoA
also makes FADH2 and NADH/H+
TCA cycle step 1
acetyl-CoA + oxaloacetate → citrate
citrate synthase
H2O → CoA-SH
condensation
rate-limiting
not reversible
TCA cycle step 2
citrate → cis-aconitate
aconitase
reversible
dehydration (H2O released)
TCA cycle step 3
cis-aconitate → isocitrate
aconitase
reversible
hydration (H2O put in)
TCA cycle step 4
isocitrate → α-ketoglutarate
isocitrate dehydrogenase
rate-limiting
not reversible
oxidative decarboxylation (CO2 released)
NADH released
manganese in the enzyme active site helps to stabilize the intermediate
TCA cycle step 5
α-ketoglutarate → succinyl-CoA
α-ketoglutarate dehydrogenase
rate-limiting
not reversible
CoA-SH → CO2
oxidative decarboxylation
NADH released
TPP, NAD, FAD co-factors
TCA cycle step 6
succinyl-CoA → succinate
succinyl-CoA synthetase
reversible
GDP/ADP → GTP/ATP
CoA-SH released
substrate-level phosphorylation
TCA cycle step 7
succinate → fumarate
succinate dehydrogenase
reversible
FADH2 released
dehydrogenation
electrons flow from FAD→Fe/S→ETC
TCA cycle step 8
fumarate → malate
fumarase
reversible
hydration (H2O put in)
TCA cycle step 9
malate → oxaloacetate
malate dehydrogenase
NADH released
dehydrogenation
reversible
pyruvate → acetyl-CoA
inhibited by ATP, acetyl-CoA, NADH, fatty acids
activated by AMP, CoA, NAD+, Ca2+
acetyl-CoA → citrate
inhibited by NADH, succinyl-CoA, citrate, ATP
activated by AMP
isocitrate → α-ketoglutarate
inhibited by ATP, NADH
activated by Ca2+, ADP
α-ketoglutarate → succinyl-CoA
inhibited by succinyl-CoA, NADH
activated by Ca2+
what does coupling depend on?
sequential redox reactions that pass e- from NADH to O2
compartmentalization of these reactions within the mitochondrion
generation of proton gradient
2 ways ATP is produed
substrate level phosphorylation
oxidative phosphorylation
electron transport
e- carried by reduced co-enzymes are passed through chain of proteins and coenzymes
drives generation of proton gradient across inner mitochondrial membrane
oxidative phosphorylation
proton gradient runs downhill to drive synthesis of ATP
standard reduction potential
Eº’
measure of how easily a compound can be reduced
more positive = the more the compound “wants” electrons
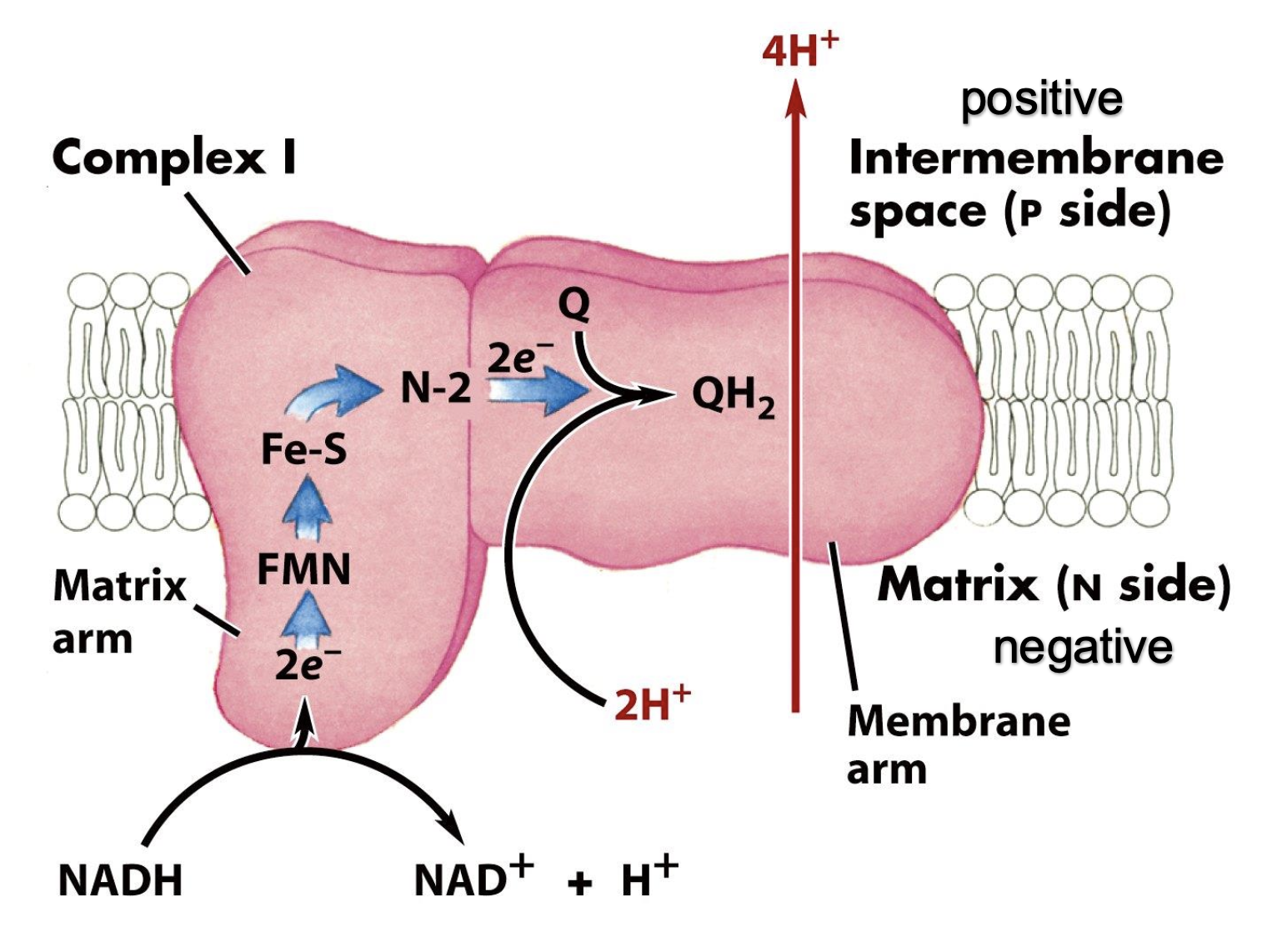
complex I prosthetic groups
FMN, Fe-S
complex II prosthetic groups
FAD, Fe-S
complex III prosthetic groups
Heme, Fe-S
complex IV prosthetic groups
Hemes; CuA, CuB
ubiquinone
lipid-soluble carrier molecule
CoQ, or Q
lives/moves in mitochondrial membrane
complete reduction requires 2e- and 2p+ (get them from matrix)
shuttle e- from complex I and II → III
complex I
NADH-dehydrogenase
FMN and Fe-S centers
electron flow: NADH — FMN, FMNH2— Fe3+, Fe3+ — Fe2+
e- ultimately shuttled to Q
energy of electron transfer used to pump 4H+