Lec 20 & 21 Enzyme Regulation
1/61
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
62 Terms
SL-3 Enzyme Regulation
Enzymatic reactions (thus products) are regulated to match ________
cell requirements
SL-3 Enzyme Regulation
achieved by regulating enzyme:
1) ______
2) _______
1) Abundance
2) Activity
SL-3 Enzyme Regulation
Abundance is controlled by ________
enzyme regulation is a natural ___________ phenomenon.
gene expression (not treated)
physiological
SL-4 Enzymatic reaction velocity
The velocity of a reaction is typically controlled by the concentrations of ______ and _______
substrates and cofactors
SL-4 Enzymatic reaction velocity
What are some cofactors?
metal ions or organic coenzymes that participate in some enzyme reactions
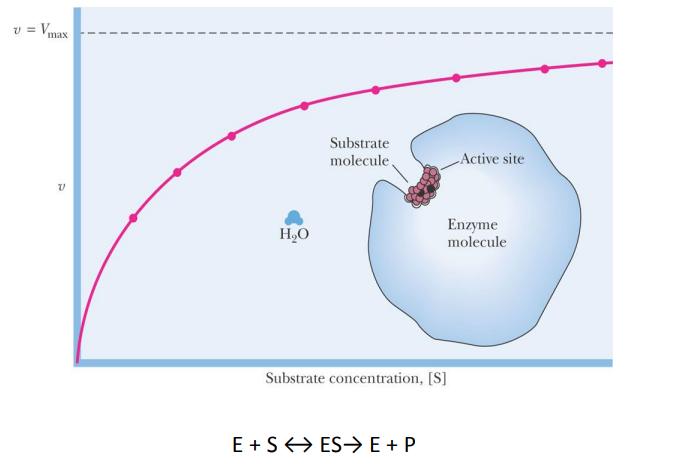
SL-5 Enzyme Regulation
Accumulation of product reduces ______ of reaction
velocity
SL-5
For conversion of substrate, S, to product, P, reaction velocity v is given by ________
d[P]/dt
SL-5
Keq = _____
Keq = [P]/[S]
SL-5
Once the ratio of [P]/[S] approaches Keq, no further reaction is apparent, because of the ___________
increased rate of the reverse reaction
SL- 5
Some enzymes may also be _______regulated by their product
allosterically
SL-6
What are the different ways to regulate an Enzyme?
1) Covalent modification
2) Zymogen (proenzyme) activation
3) Isozymes
4) Control by modulatory proteins
5) Allosterically
SL-7 Enzyme Regulation – covalent modification
Covalent modification of an amino acid side chain can _______ an enzyme (e.g. : Serine, Threonine, Tyrosine, Aspartate)
Covalent modification of an amino acid side chain can activate or inactivate an enzyme (e.g. : Serine, Threonine, Tyrosine, Aspartate)
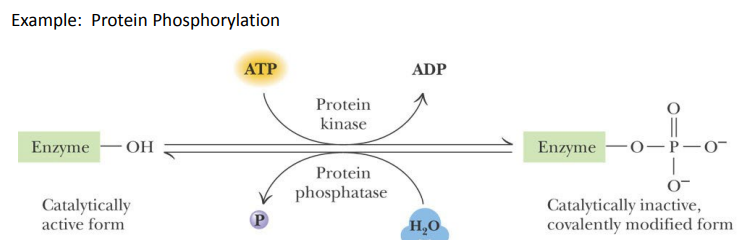
SL-7
The enzymes that introduce and remove modifications can be ________
regulated (e.g. by allosteric control or by covalent modification)
SL-7
How can we control the generation of Products?
By controlling the ratios between inactive /active enzymes the generation of products can be controlled
SL-8 Enzyme Regulation – covalent phosphorylation
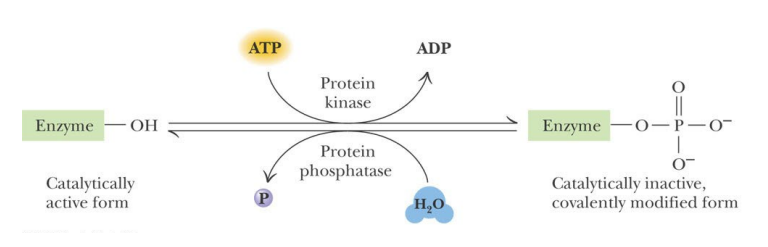
SL-8 Enzyme Regulation – covalent phosphorylation
What is the most prominent form of covalent modification in cellular regulation?
Reversible phosphorylation
SL-8 Enzyme Regulation – covalent phosphorylation
Phosphorylation is accomplished by _________
Phosphorylation is accomplished by protein kinases
SL-8 Enzyme Regulation – covalent phosphorylation
Each __________ targets specific proteins for phosphorylation
Each protein kinase targets specific proteins for phosphorylation
SL-8 Enzyme Regulation – covalent phosphorylation
Phosphoprotein phosphatases catalyze the _________
Phosphoprotein phosphatases catalyze the reverse reaction – removing phosphoryl groups
SL-8 Enzyme Regulation – covalent phosphorylation
________ and ____________ themselves are targets of regulation
Kinases and phosphatases themselves are targets of regulation
SL-9 Other Covalent Modification that Regulates Protein Function
Several different chemical modifications of proteins have been discovered
Only a few are used to achieve __________ regulation through __________ of an enzyme between active and inactive forms
Several different chemical modifications of proteins have been discovered
Only a few are used to achieve metabolic regulation through reversible conversion of an enzyme between active and inactive forms
SL-10
_________ is a prominent modification for the regulation of metabolic enzymes
Acetylation
SL-10
Acetylation of an _______ group on a _________ residue changes it from a ______ charged amino group to a _______
Acetylation of an ε-NH3 + group on a Lys residue changes it from a positively charged amino group to a neutral amide
SL-10
acetylation - This change may have consequences for __________ and thus _______
This change may have consequences for protein structure and thus function
SL-10
The acetylating enzyme is termed an _________ or ________
The acetylating enzyme is termed an acetyl-CoA-dependent lysine acetyltransferase or KAT (More than 30 KATs are known in mammals)
SL-10
___________ by KDACs (lysine deacetylases) reverse the effects
Deacetylation by KDACs (lysine deacetylases) reverse the effects
SL-10
Acetylation of metabolic enzymes is an important mechanism for ____________
Acetylation of metabolic enzymes is an important mechanism for regulating the flow of metabolic substrates (e.g: carbohydrates and fats)
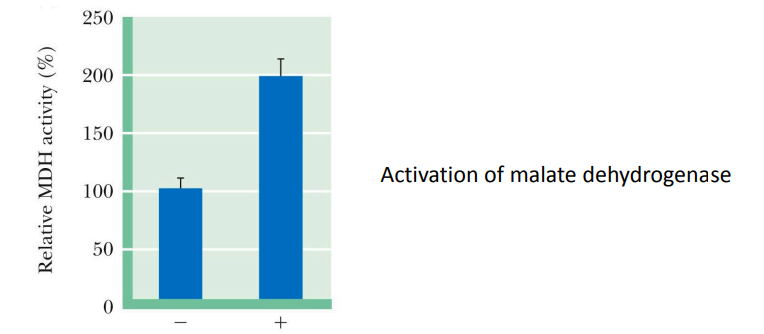
SL-11
Synthesis of enzyme as an inactive precursor Activation requires specific __________
proteolytic cleavage
SL-11
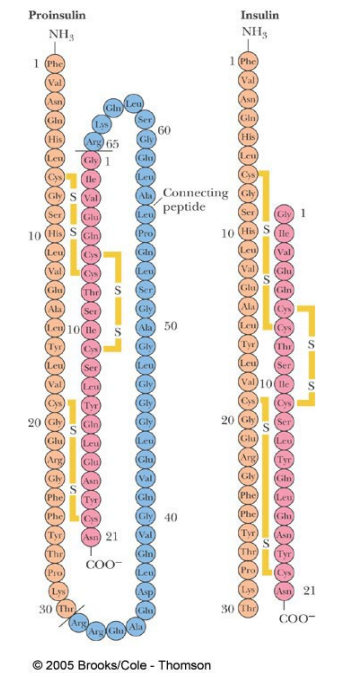
The hormone insulin (responsible for glucose uptake) is made as an inactive _________ precursor (proinsulin).
Proteolytic removal of __________ generates the active form, (two chains and three disulfide bonds)
The hormone insulin (responsible for glucose uptake) is made as an inactive 86 amino acid precursor (proinsulin).
Proteolytic removal of residues 31-65 generates the active form, (two chains and three disulfide bonds)
SL-11
Proenzyme activation by protease cleavage is _________ (reversible / irreversible)
Proenzyme activation by protease cleavage is irreversible
SL-12 Proenzymes of the digestive tract
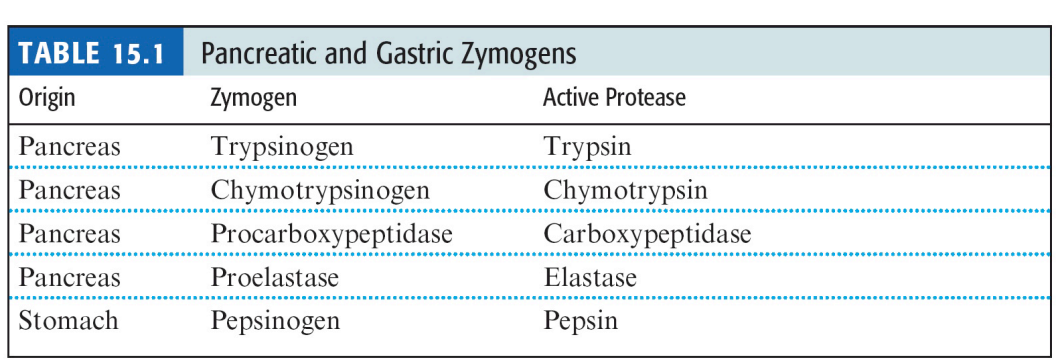
SL-13 Proenzymes of the digestive tract
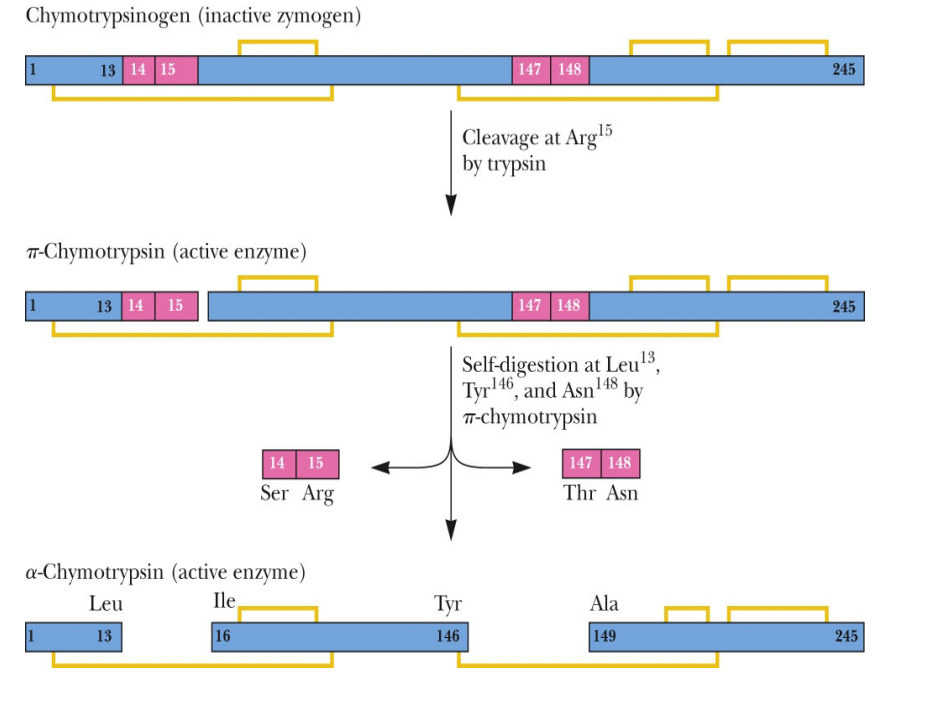
SL-14 Enzyme regulation - Isozymes
____________ equivalent subunits (small primary sequence differences not identical) but _________ distinct subunits
Structurally equivalent subunits (small primary sequence differences not identical) but catalytically distinct subunits
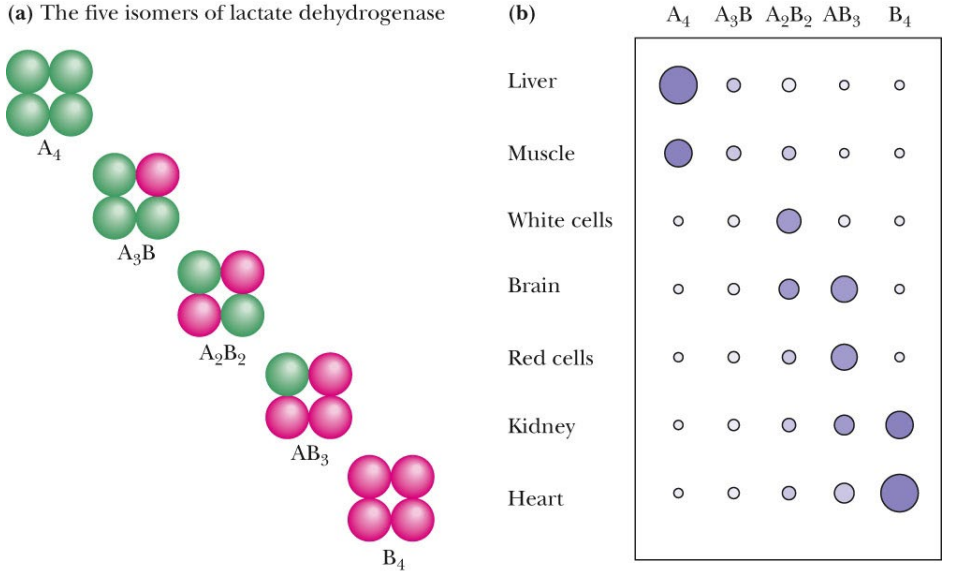
SL-14 Enzyme regulation - Isozymes
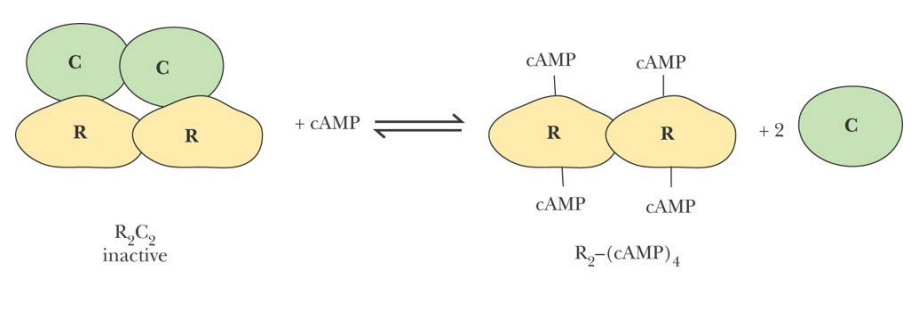
SL-15 Enzyme regulation – Binding of regulatory proteins
The catalytic (C) subunit of cAMP-dependent protein kinase is kept in an _______ form by the _______ subunit. Binding of c-AMP to the __ subunit releases the active enzyme.
The catalytic (C) subunit of cAMP-dependent protein kinase is kept in an inactive form by the regulatory (R) subunit. Binding of c-AMP to the R subunit releases the active enzyme.
SL-16 Allosteric regulation
What is Allosteric regulation?
Activation or inhibition of enzyme activity by small molecules (metabolites) that interact non-covalently with the enzyme
SL-16
The ________ binds to a site other than the substrate binding site (allo = other)
The allosteric effector binds to a site other than the substrate binding site (allo = other)
SL-16
Reversible binding of an effector to the enzyme allows for __________ and thus _______ of enzymatic activity
Reversible binding of an effector to the enzyme allows for very rapid response times and thus rapid control of enzymatic activity
SL-17 Allosteric regulation
Example: the product (F) as an allosteric inhibitor of the first enzyme of the pathway (E1).
This is called ________
E1 is called _________
Allosteric ________ is also common
feedback inhibition
E1 is called regulatory enzyme: Where activity of the metabolic pathway is regulated
Allosteric activation is also common
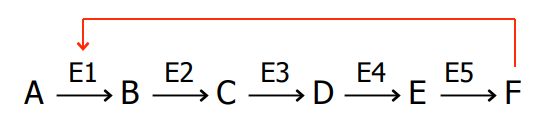
SL-18
Regulatory enzymes are often subject to _______ regulation
allosteric
SL-18 Allosteric regulation – regulatory enzymes
Kinetic does not follow the _________ equation
Michaelis-Menten
SL-18 Allosteric regulation – regulatory enzymes
Properties of regulatory enzymes:
________ kinetics
Often show _______
_______ substrate binding is a special case of allostery
Regulatory enzymes are _______
Sigmoid/S-shaped kinetics
Often show Cooperativity: Binding of S make binding of other S easier to same molecules
Cooperative substrate binding is a special case of allostery
Regulatory enzymes are oligomeric (it follows from cooperative kinetics, more than one substrate binding site)
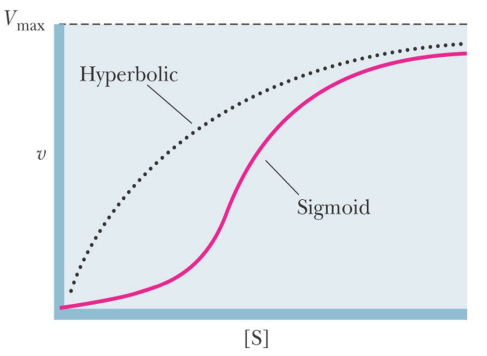
SL-19 Allosteric regulation – regulatory enzymes
Properties:
Inhibition by _________ inhibitor does not conform to normal inhibition patterns
Allosteric effector (inhibitor or activator) usually has _____ structural similarity to the substrate
Effector binds at a site ______ from the substrate binding site
Regulatory effects alters distribution of ______ distribution and __________ changes occurring in the enzyme as a result of _________
Inhibition by feedback allosteric inhibitor does not conform to normal inhibition patterns
Allosteric effector (inhibitor or activator) usually has little/no structural similarity to the substrate
Effector binds at a site remote from the substrate binding site
Regulatory effects alters distribution of enzyme distribution and conformational changes occurring in the enzyme as a result of effector binding
SL-20 Symmetry model for allosteric regulation: the MonodWyman-Changeux (MWC) model
what are the 2 states of allosteric proteins?
Allosteric proteins can exist in two states: R (relaxed) and T (taut)
SL-20
In the MWC Model , all the subunits of an oligomer must be in the _________ state
In the MWC Model , all the subunits of an oligomer must be in the same state
SL-20 Symmetry model for allosteric regulation: the MonodWyman-Changeux (MWC) model
T state predominates in the absence of __________
S binds much tighter to ___ than to ____
In the absence of ligand, the two states are called _____ and ____
The equilibrium constant (L) for the T0/R0 equilibrium is ______ , that is T0 predominates
T state predominates in the absence of substrate S
S binds much tighter to R than to T
In the absence of ligand, the two states are called R0 and T0
The equilibrium constant (L) for the T0/R0 equilibrium is large, that is T0 predominates
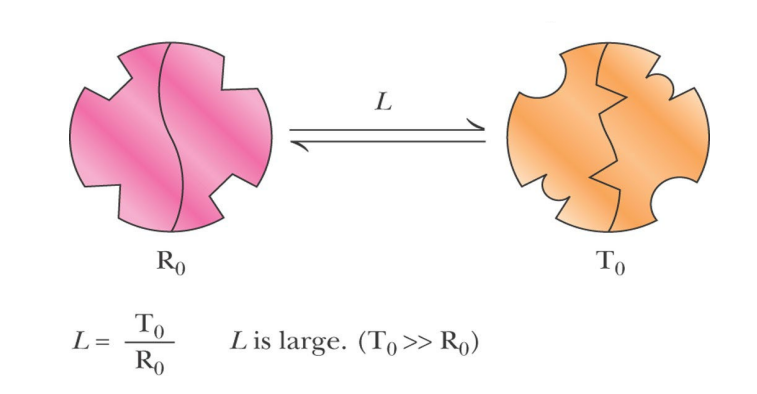
SL-20 Symmetry model for allosteric regulation: the MonodWyman-Changeux (MWC) model
The substrate affinity of each state of the enzyme is described by a ____________ constant: _________(for the relaxed form) and __________ (for the tense form)
KT is much ______(smaller/greater) than KR. That is, R0 has a higher affinity for the substrate than T0
The substrate binds with higher affinity to the less abundant form of the enzyme
The substrate affinity of each state of the enzyme is described by a dissociation constant: KR (for the relaxed form) and KT (for the tense form)
KT is much greater than KR. That is, R0 has a higher affinity for the substrate than T0
The substrate binds with higher affinity to the less abundant form of the enzyme
SL-22 MWC Model - cooperativity
Substrate binds preferentially to R0 . The substrate bound form is R1 •
Binding lowers the abundance of R0
Equilibrium between T0 to R0 conformationsis driven to R0
R has >1 substrate binding sites
Substrate binding increases the concentration of R (i.e. R1 + R0) •
Therefore, the amount (number) of substrate binding sites increases : positive cooperativity
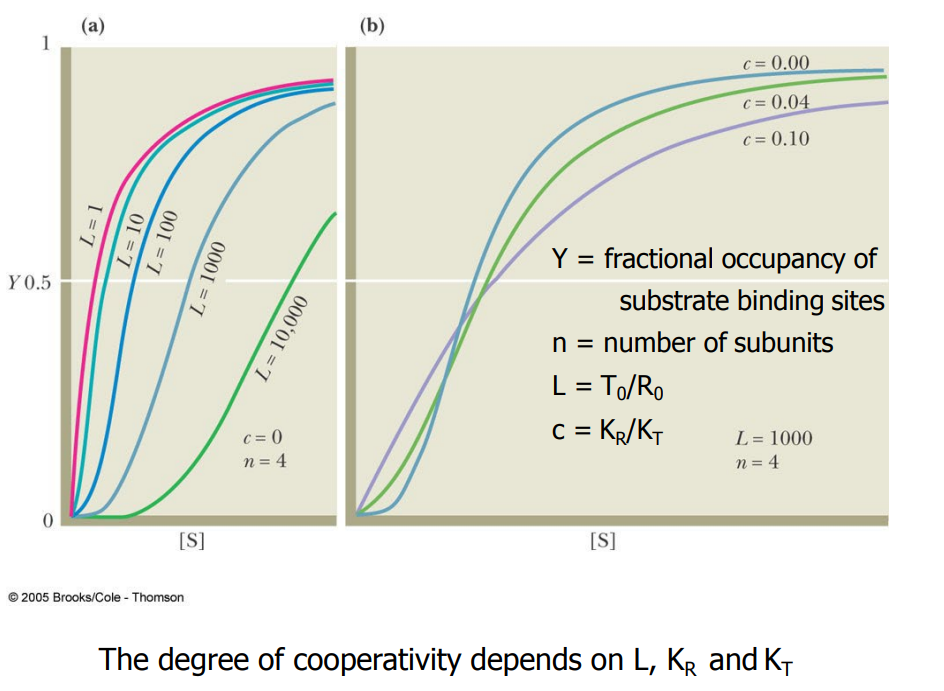
SL-23 MWC Model cooperativity
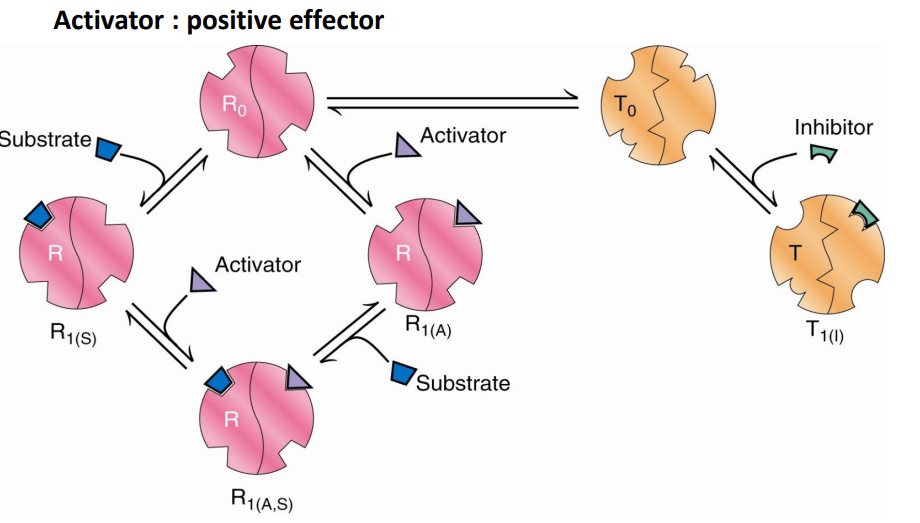
SL-24 The Monod-Wyman-Changeux (MWC) model : allosteric regulation
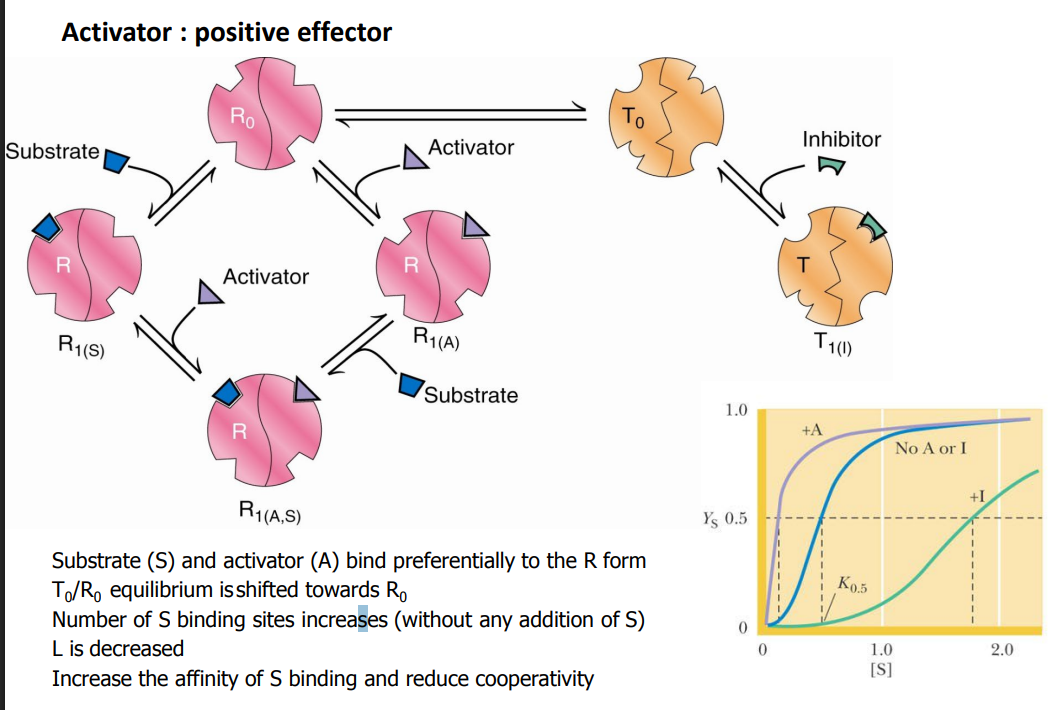
Activator : positive effector
_______ and _______ bind preferentially to the __ form
_______ equilibrium is shifted towards ___
Number of S binding sites _______(increases/decreases)
L is _______ (increased/decreased)
_______ the affinity of S binding and reduce _______
Substrate (S) and activator (A) bind preferentially to the R form
T0/R0 equilibrium is shifted towards R0
Number of S binding sites increases (without any addition of S)
L is decreased
Increase the affinity of S binding and reduce cooperativity
SL-25 Quiz
In the presence of a positive effector the YS/[S] curve will:
A - Shift to the right
B - Shift to the left
C - Shift vertically
D – None of the above
B
SL-26 The Monod-Wyman-Changeux (MWC) model : allosteric regulation
Activator : positive effector
Substrate (S) and activator (A) bind preferentially to the R form
T0/R0 equilibrium is shifted towards R0
Number of S binding sites increases (without any addition of S)
L is decreased
Increase the affinity of S binding and reduce cooperativity
SL-27 The Monod-Wyman-Changeux (MWC) model : allosteric regulation
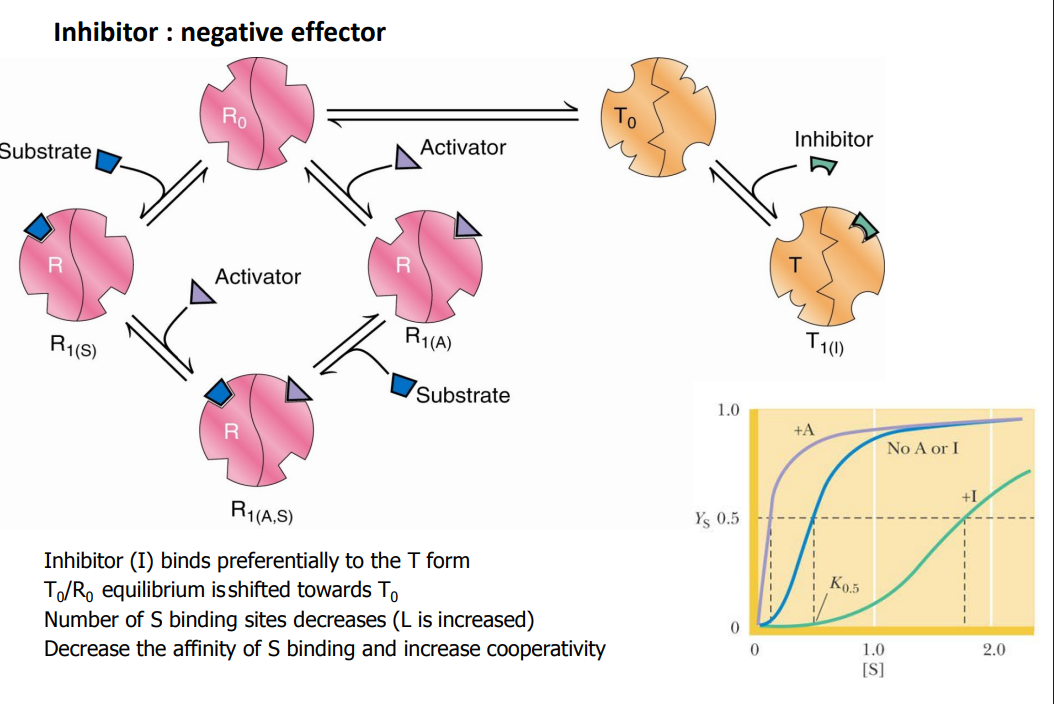
Inhibitor : negative effector
_________ binds preferentially to the ___ form
T0/R0 equilibrium is shifted towards _____
Number of S binding sites _______
L is _______
_______ the affinity of S binding and ________ cooperativity
Inhibitor (I) binds preferentially to the T form
T0/R0 equilibrium is shifted towards T0
Number of S binding sites decreases (L is increased)
Decrease the affinity of S binding and increase cooperativity
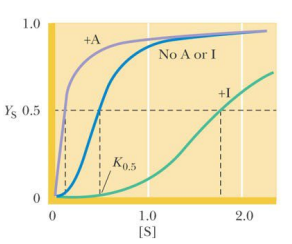
SL-28 The Monod-Wyman-Changeux (MWC) model: allosteric regulation – K systems and V systems
‘K systems’:
the concentration of substrate that gives half-maximal substrate binding (K0.5) changes in the presence of A and I.
Vmax does not change
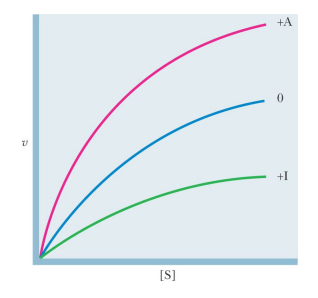
SL-28 The Monod-Wyman-Changeux (MWC) model: allosteric regulation – K systems and V systems
‘V systems’
In ‘V systems’, allosteric effectors change the Vmax and K0.5 remains the same
R and T forms of the enzyme have the same affinities for S, but differ in their affinities for A and I and in their catalytic properties
This regulation is important when the cellular concentration of S is much greater that K0.5
SL-29 The Monod-Wyman-Changeux (MWC) model : allosteric regulation
In the case of positive cooperativity,
the substrate acting as a ______ effector, in this case called a ________
In the case of positive cooperativity,
the substrate acting as a positive effector, in this case called a positive homotropic effector
SL-29 The Monod-Wyman-Changeux (MWC) model : allosteric regulation
A ligand other than the substrate that activates the binding of substrate is called a ________ effector or allosteric _______
A ligand other than the substrate that activates the binding of substrate is called a positive heterotropic effector or allosteric activator
SL-29 The Monod-Wyman-Changeux (MWC) model : allosteric regulation
A ligand that Inhibit the binding of substrate is called a ________ effector or allosteric _______
A ligand that Inhibit the binding of substrate is called a negative heterotropic effector or allosteric inhibitor
SL-29 The Monod-Wyman-Changeux (MWC) model : allosteric regulation
The MWC model cannot explain _______ in substrate binding
The MWC model cannot explain negative cooperativity in substrate binding
SL-30 The Koshland-Nemethy-Filmer (KNF) model : allosteric regulation
There is no equilibrium between different _________ of the enzyme with no ____
S binding ______ a ________ change, and the subunits can adopt different ______
S binding causes the other subunit to undergo a ____ change, to a form that has a higher or lower ____ for S. The model can explain________ cooperativity
_______ activator works in the same way as S, but by binding to a different site
_________ works by preventing the __________ change induced by ____ binding
There is no equilibrium between different conformers of the enzyme with no S
S binding causes a conformational change, and the subunits can adopt different conformations (unlike in MWC)
S binding causes the other subunit to undergo a conformational change, to a form that has a higher or lower affinity for S. The model can explain both negative and positive cooperativity
Allosteric activator works in the same way as S, but by binding to a different site
Inhibitor works by preventing the conformational change induced by S binding
SL-31 Key differences between the MWC and KNF models cooperativity
MWC
In MWC, there is a pre-existing equilibrium between ___ and ___ forms, in the absence of _____.
Substrate or activator binds preferentially to the ___ form
All subunits must be in the ______ conformation, there are ____(number) possible conformations
All subunits change conformation _____, therefore mechanism is called the ______ or _______ model
In MWC, there is a pre-exisiting equilibrium between R and T forms, in the absence of ligand.
Substrate or activator binds preferentially to the R form
All subunits must be in the same conformation, there are only 2 possible conformations All subunits change conformation together, therefore mechanism is called the concerted or symmetry model
SL-31 Key differences between the MWC and KNF models cooperativity
KNF
In KNF, conformational change is induced by _________
Conformational change can be transmitted to a _________ subunit
__________ conformations are possible
Subunits can be in _________ (same/different) conformations and change ___________, so mechanism is called sequential model
In KNF, conformational change is induced by ligand binding
Conformational change can be transmitted to a neighboring subunit
Intermediate conformations are possible
Subunits can be in different conformations and change sequentially, so mechanism is called sequential model