DNA Replication, Repair Biochem
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
21 Terms
DNA Replication Description
DNA replication is semiconservative.
The two DNA strands separate and each is copied so that the new DNA double helices contain one original strand and one newly synthesized strand.
DNA replication is bidirectional.
Replication proceeds in both directions from origin of replication
Replication is semidiscontinuous
One of the two strands is copied discontinuously as a set of fragments called Okazaki fragments which are then joined to form an intact shard.
Okazaki fragments are 1000-2000 nucleotides long in E. coli, 100-200 in eukaryotes.
Examples
Bidirectional replication separates strands, brings in nucleotides to make new strands (replication origin).
Unidirectional means only left side is unwound to make new strands.
Bidirectional unwinds both sides.
E. coli only has 1 replication origin (bidirectional replication) called OriC'
Humans have 30,000 replication origins
For semi-discontinuous replication, leading strand is synthesized by DNA polymerase, and Okazaki fragments also joined to form lagging strand as well. 5’→3’ end only.

Semi-Discontinuous DNA Replication Due To:
DNA polymerases only synthesize DNA 5’→3’
The two DNA strands of the double helix must be in opposite directions.
One strand 5’→3’, the other 3’→5’
Mechanism of DNA Replication (Initialtion)
In the bacterium E. coli, DNA replication consists of initiation, elongation, and termination phases.
Initiation
Replication is initiated at the origin of replication (oriC) by the binding of DnaA protein (the initiation factor), which separates the strands of the double helix.
DnaB protein (a helicase, or DNA-unwinding protein) binds to the separated strands and moves along the DNA, unwinding the double helix as it moves.
SSB (single-stranded DNA binding protein) prevents the two strands from coming back together.
DNA gyrase (DNA topoisomerase II) relieves stress in the helical DNA structure caused by DNA unwinding.
Initiation of DNA Replication
DnaA binds to origin of replication, forming a bead.
The DnaA bead locally unwinds the the DNA into replication forks.
2 DnaB (circular 6 subunits) transported to unwound area by DnaC and DnaT. Unwinds the strands in bidirectional manner.
DnaB continues to move along the helix and separate strands, while SSB (tetramers) bind to unwound bases so they cannot base-pair.

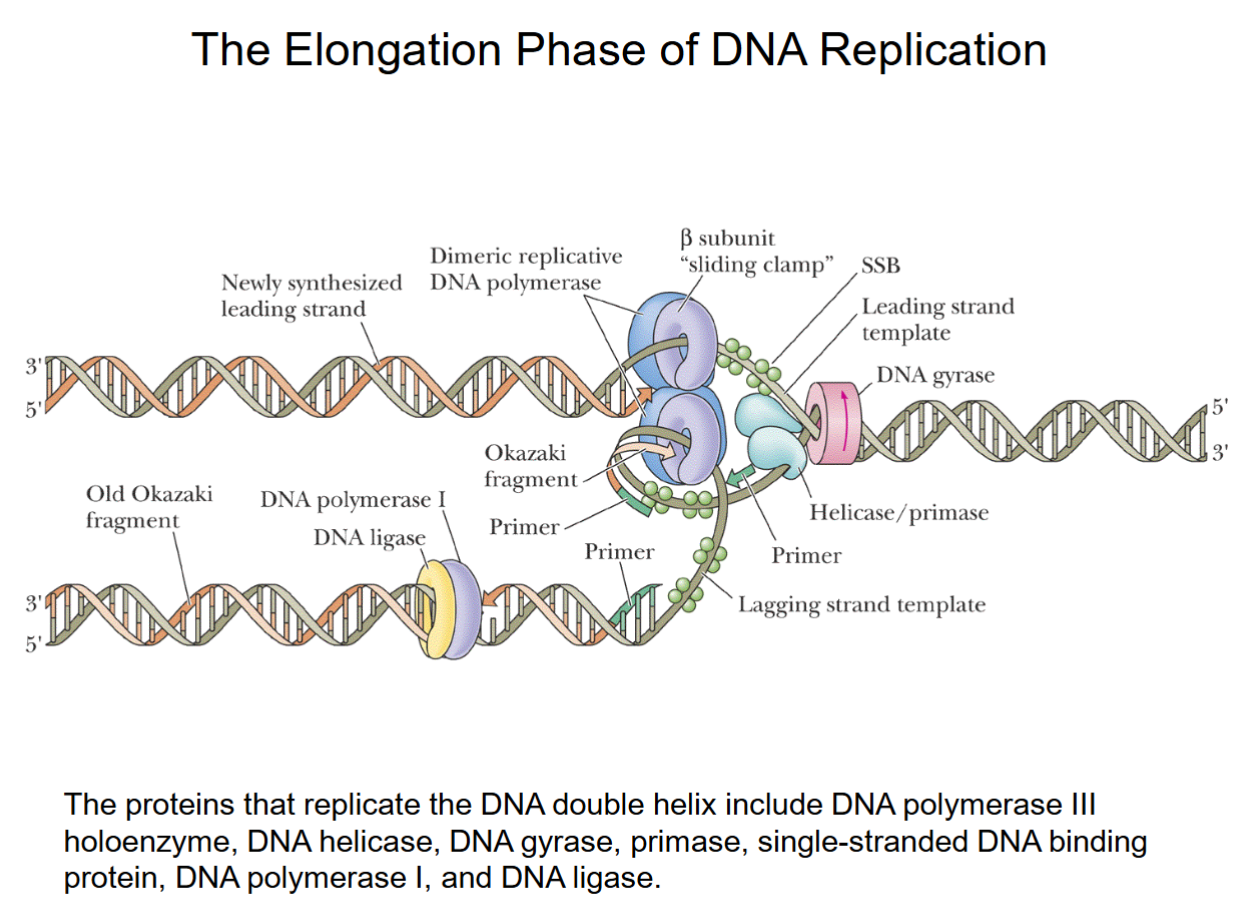
Mechanism of DNA Replication (Elongation)
The proteins at the replication fork that carry out the elongation phase of DNA replication include:
(a) DNA unwinding proteins (DNA helicase (DnaB), gyrase).
(b) Primase enzyme, which synthesizes RNA primer needed for DNA replication.
(c) DNA polymerase III holoenzyme, which synthesizes new DNA strands.
DNA polymerase does NOT know how to start a chain, needs a 3’ end with an -OH from RNA primer.
The Elongation Phase of DNA Replication
DnaB helicase/gyrase work together.
Primase produces primers for DNA polymerase to start synthesis (every strand must begin with a primer).
SSB tetramers bound temporarily to prevent strands recombining.
One part of dimeric replicative DNA polymerase (III holoenzyme) continuously synthesizes the top leading strand from 1 primer (5-10 nucleotide length).
Okazaki fragments produced in lower lagging strand from >1000 primers
DNA polymerase I removes primers from lagging strand, synthesizes short patch to fill where primers were. DNA ligase joins it to the strand.
After this, lagging strand is fully formed.
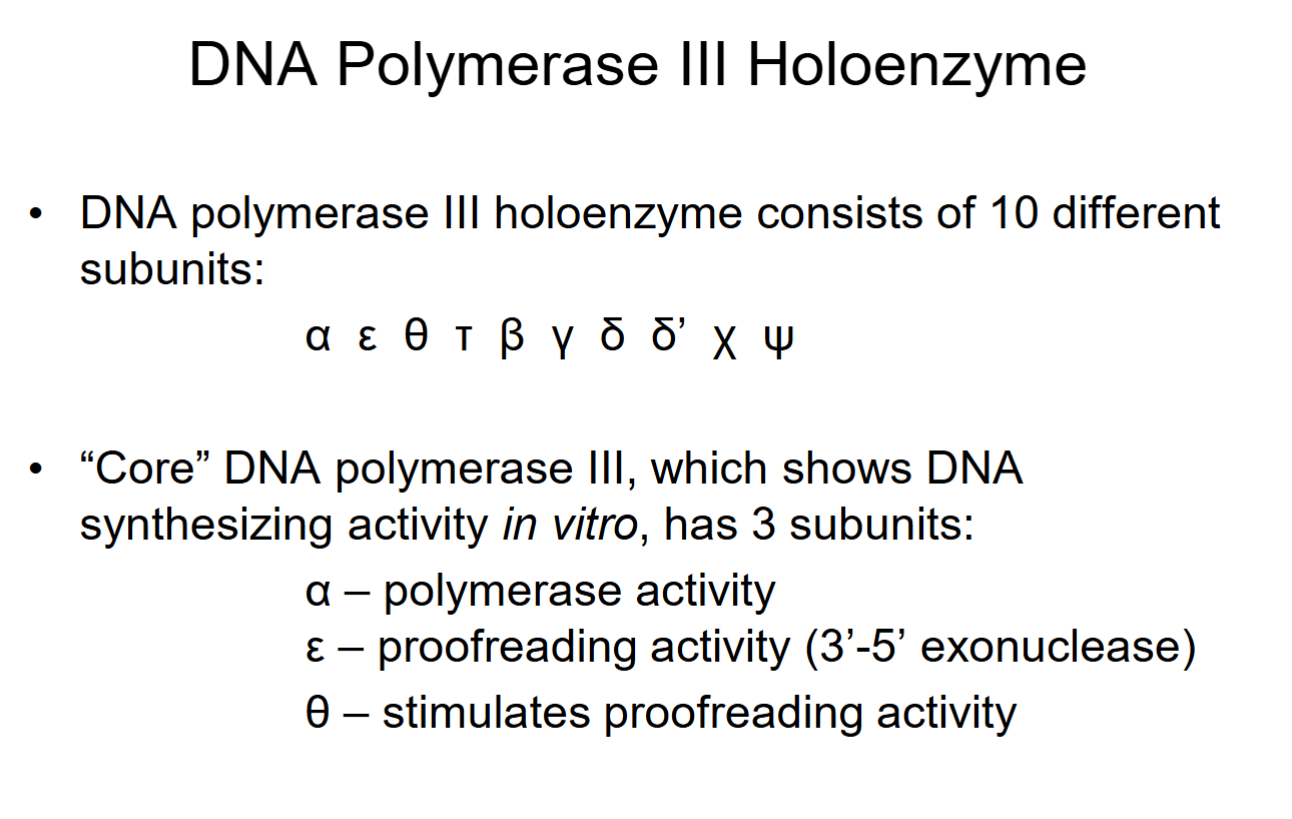
DNA Polymerase III Holoenzyme
Polymerase synthesizes from 5’→3’ end, so 3’ end is constantly changing as nucleotides are added.
Mistakes spotted by ε/θ, improper base-pairings cause process to stop, back up, and and fix it.
3’-5’ exonuclease moves 3’ end back 1 nucleotide in 3’→5’ direction to cleave off the end (with the
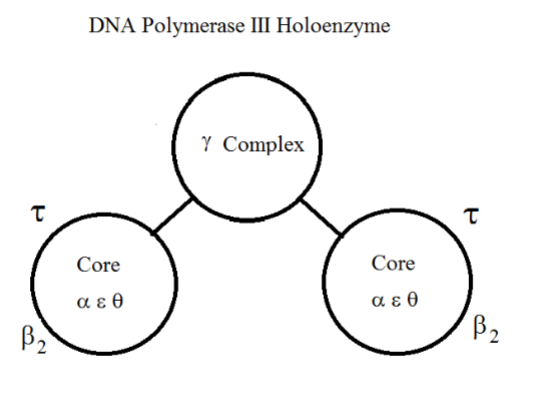
DNA Polymerase III Holoenzyme
2 β-dimer subunits keep core bound to DNA.
Gamma, delta, delta’, chi (x), psi subunits in the gamma complex to bridge both cores.
Mechanism of DNA Replication (Termination)
The two bidirectional replication forks continue to move around the circular E. coli chromosome until they meet at a termination region (Ter region), and replication is terminated.
During termination, a replication termination protein, Tus, is bound to Ter and helps bring about termination.
DNA Replication in Eukaryotic Cells
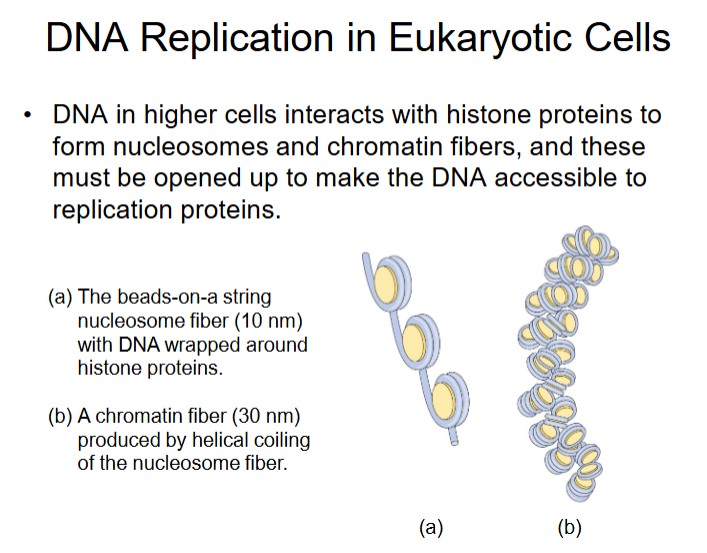
Eukaryotic DNA Polymerases
Polymerase α: functions in the initiation of nuclear DNA replication.
Polymerase δ: principal DNA polymerase in eukaryotic DNA replication
Polymerase ε: cooperates with DNA polymerase δ.
Polymerase β: functions in DNA repair (removes damages segments and replaces with functional DNA).
Polymerase γ: DNA replicating enzyme of mitochondria.
Polymerase α consists of RNA and DNA synthesizing activity (DNA only enough to start process). Made up of POLA1 (catalytic subunit) and POLA2 for DNA polymerase activity as well as PRIM1/PRIM2 for primase subunits.
Polymerase δ does most of the DNA replication. Made up of POLD1(catalytic subunit)/2/3/4 subunits and PNCA which is a DNA clamp. Catalytic subunits have the actual DNA synthesizing activity.
Biochemical Properties of the Principal Eukaryotic DNA Polymerases

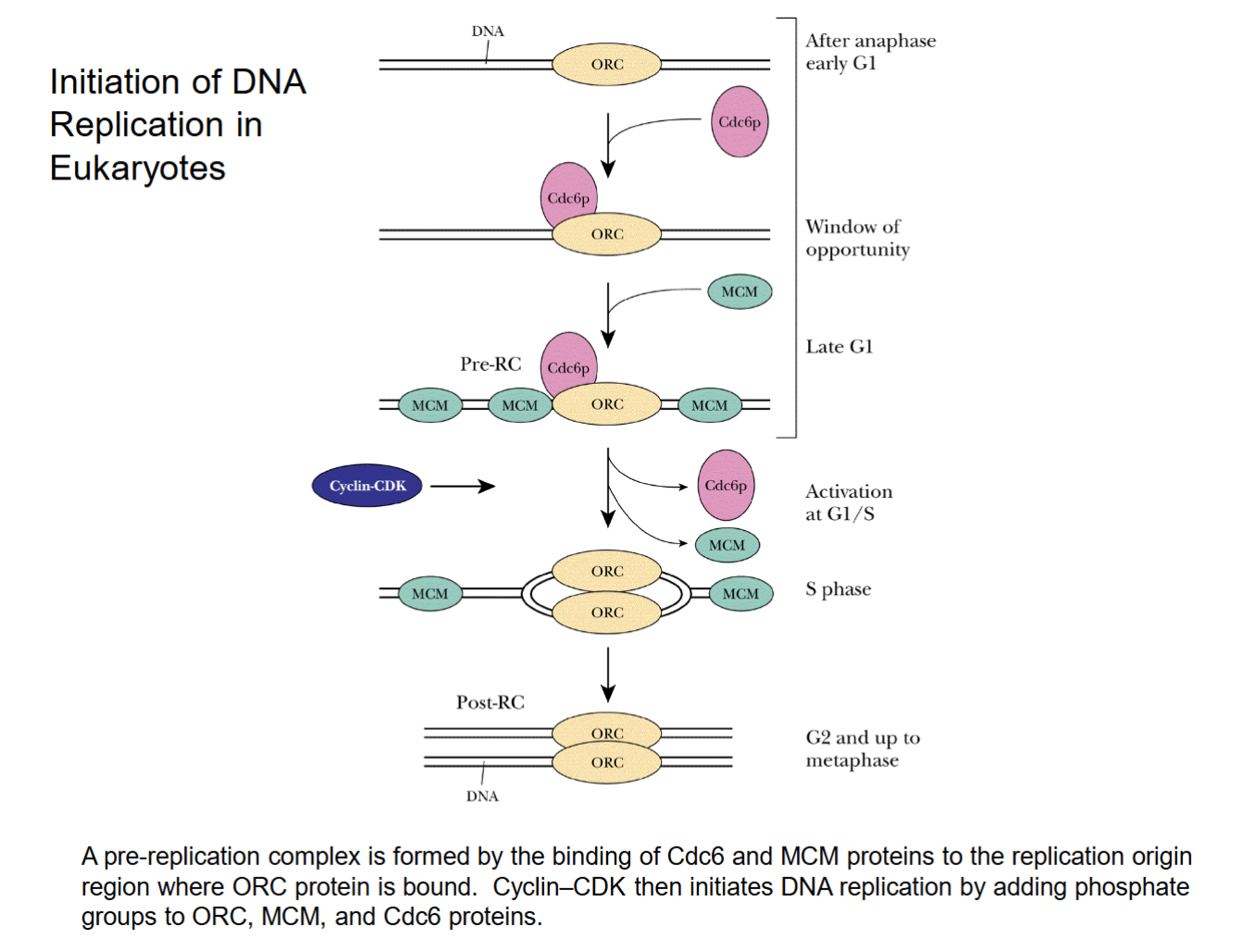
Initiation of DNA Replication in Eukaryotes
ORC: protein complex that binds to replication origins; essential for assembly of the pre-replicative complex
MCM proteins: eukaryotic helicases that unwind the double helix at replication origins and separate the DNA strands (does the job of DnaA and DnaB in E. coli.)
Cdc6 proteins: bring about the binding of MCM proteins to replication origins
Cyclin CDK: kinase complex that initiates DNA replication by adding phosphate groups to ORC, MCM, and Cdc6 proteins.
Changes net charge, which changes folding. Actives proteins by changing conformation, especially MCM.
Initiation of DNA Replication in Eukaryotes
Cdc6 brings in MCM to ORC site.
Cyclin CDK removes Cdc6 and some MCM, causes strands to separate as MCM is activated. Turns into “S phase”, MCM splits apart strands.
No termination site/proteins. Replication forks merge across strand until fully separated.
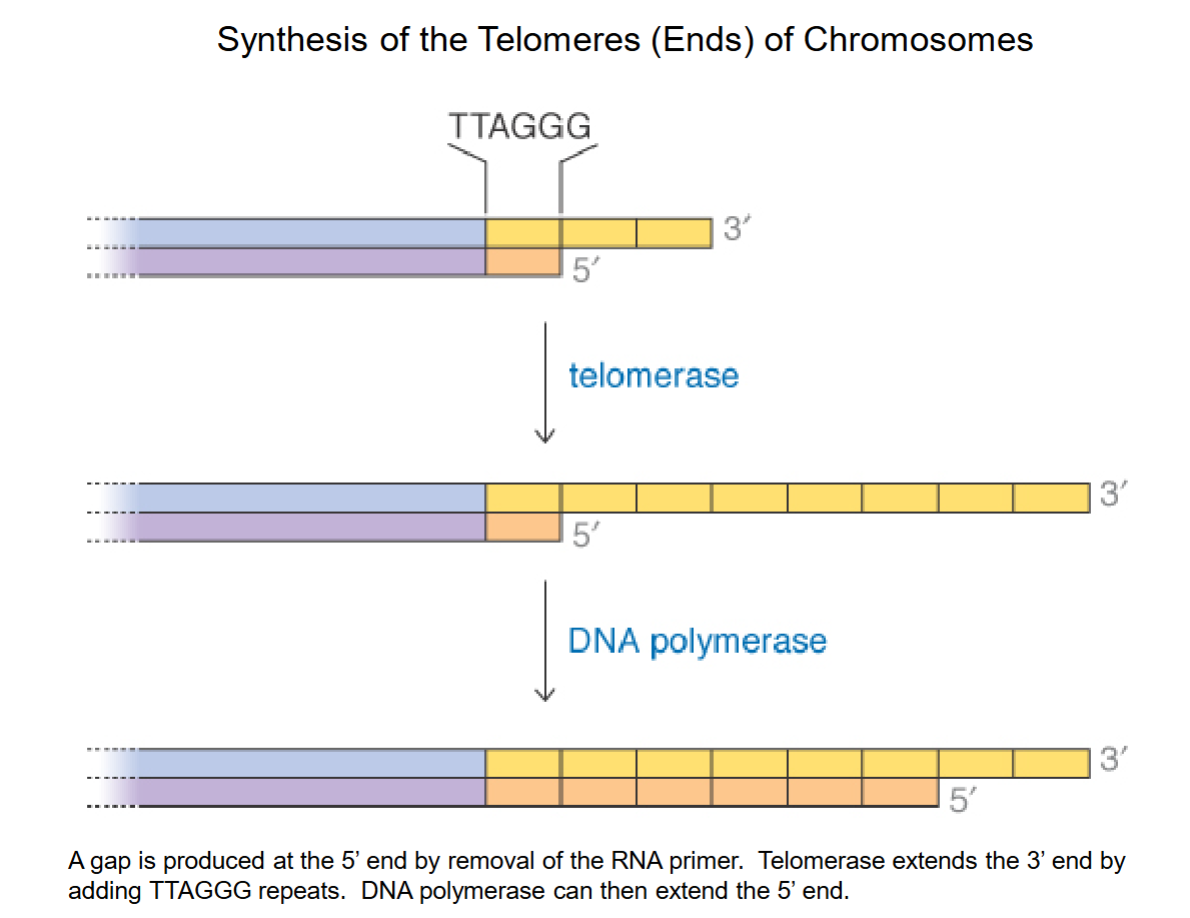
Synthesis of Telomeres (Ends) of Chromosomes
PROBLEM: 5’ gap where primer used to be! Single stranded DNA is fragile, easily broken by DNA nuclease, but is not viable to shorten DNA in the long run.
Telomerase extends 3’ end w/ 6-base repeat of TTAGGG.
DNA polymerase can now bind to longer stretch (was too short before) to produce complimentary sequences and extend the 5’ end.
A special adaption in eukaryotic linear DNA to prevent losing DNA.
Adults have ~3000 extra bp, ~8000 in newborns.
DNA Repair
DNA damage can result from environmental factors or from errors during DNA synthesis.
The most common forms of damage are:
(a) A chemically damaged, incorrect, or missing base.
(b) Longer damaged DNA segments; Deletions or insertions.
(c) Pyrimidine dimers (adjacent pyrimidine bases such as thymine, uracil, cytosine can be covalently linked by UV light)
(d) DNA single-strand or double strand breaks.
(e) Covalent cross-linking of strands.
Because of its importance, DNA is the only molecule that is repaired by the cell. About 10,000 bases per cell are lost each day by damage.
Molecular Mechanisms of DNA Repair
Cells have a number of systems to repair the different types of DNA damage:
Mismatch repair system
Corrects errors introduced when DNA is replicated
Base excision repair
Acts on single bases damaged by oxidation or other chemical modifications.
Nucleotide excision repair
Repairs larger regions of damaged DNA than base excision repair (~10-20bp).
Rejoins cut double-strands:
Nonhomologous end-joining
DNA double-strand breaks caused by radiation caused by radiation or free radicals are repaired.
Recombination repair
Uses homologous recombination to repair double-strand breaks that occur during the cell-division cycle.
Direct reversal repair systems
Chemically modified bases are directly repaired; an example is photolyase, an enzyme that repairs thymine dimers.
Molecular Mechanisms of DNA Repair
Mismatch repairL The E. coli methyl-directed pathway of mismatch repair.
DNA replication is very accurate, but occasionally DNA polymerase introduces the wrong base into a newly synthesized strand, producing mismatched base pairs such as G/T or A/C.
To identify the strand with the incorrect base, mismatch repair makes use of the fact that, after replication, methyl groups (-CH3) are added to a new DNA strand.
But for a short time, only the older strand has methyl groups.
The strand without the methyl groups is the newly synthesized strand and is, by definition, the strand with the incorrect base.
An endonuclease, an exonuclease, DNA polymerase III, and DNA ligase then remove the incorrect nucleotide and replace it with a correct nucleotide.
Accurate replication of DNA is produced by:
High fidelity of DNA polymerase
Error rate = 1 in 10 million nucleotides
Proofreading activity of the polymerase
Mismatch repair system
Error rate after (2) and (3) = 1 in one billion
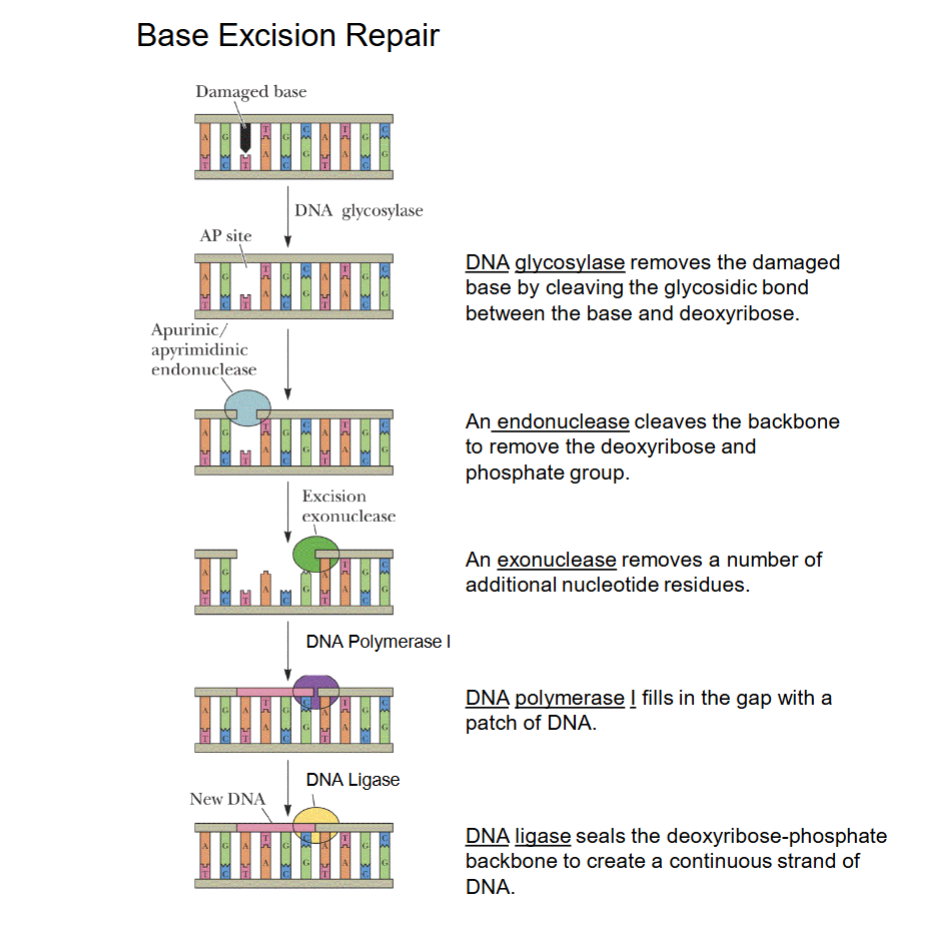
Base Excision Repair
DNA glycosylase first removes damaged base.
Endonuclease attacks DNA from inside (middle) to remove backbone segment.
Exonuclease attacks ends formed to remove additional nucleotides.
DNA polymerase I repairs patch of DNA (same enzyme in DNA replication to repair primer patches).
DNA ligase seals backbone to join nucleotides together.
Nucleotide excision repair works similarly.