Eukaryotic cell structure
1/103
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
104 Terms
What are the limits to prokaryotic cell size?
surface area : volume - lower SA per volume as gets bigger
diffusion rates of molecules
high concentrations of compounds + enzymes
What does compartmentalisation allow?
allows enzymes + substrates to be localised + concentrated
allows cells to differentiate + specialise into specific functions
What is the function of the plasma membrane?
Cell boundary. Selectively permeable with transport systems that allow cell-cell interactions, adhesion, secretion + signalling.
What is the function of the cytoplasm?
Composed of cytosol and organelles. Location of many metabolic processes
What is the function of the cytoskeleton?
Composed of actin filaments, intermediate filaments + microtubules. Provides cell structure + movement
What is the function of the endoplasmic reticulum?
transport of materials + lipid synthesis
What is the function of ribosomes?
protein synthesis
What is the function of golgi apparatus?
packaging and secretion of materials
lysosome formation
protein modification
protein sorting for movement around cell
What is the function of lysosomes?
intracellular digestion
What is the function of mitochondria?
Energy production
What is the function of chloroplasts?
photosynthesis
found in plants, algae + photosynthetic bacteria
light + CO2 → organic molecule
What is the function of nucleus?
houses genetic information
What is the function of nucleolus?
ribosomal RNA synthesis + ribosome construction
What is the function of cell wall?
strength + shape of cells
What is the function of cilia + flagella?
cell movement
What is the role of vacuole?
temporary storage, digestion + water balance
large - fluid filled vesicles
contain hydrolytic enzymes
30-90% of cell volume
What is a compartment without membranes called?
biomolecular condensates
based on selective aggregation of some macromolecules
concentrated zones of enzymes
How are biomolecular condensates formed?
requires scaffolds e.g. nucleic acid or proteins
scaffolds form multiple weak, fluctuating binding interactions with themselves (aggregate)
recruit specific proteins + nucleic acids into condensate (client proteins)
What are the characteristics of biomolecular condensates?
highly dynamic + reversible properties
liquid-like behaviour
condensates of different properties can coexist
liquid-liquid phase separation
How are cells studied?
microscopes - live imaging
fluorescent proteins - bind GFP to protein of interest to track it
What are the similarities between plant + animal cells?
membrane-bound organelles
similar membrane, cytosol + cytoskeleton elements
function of organelles
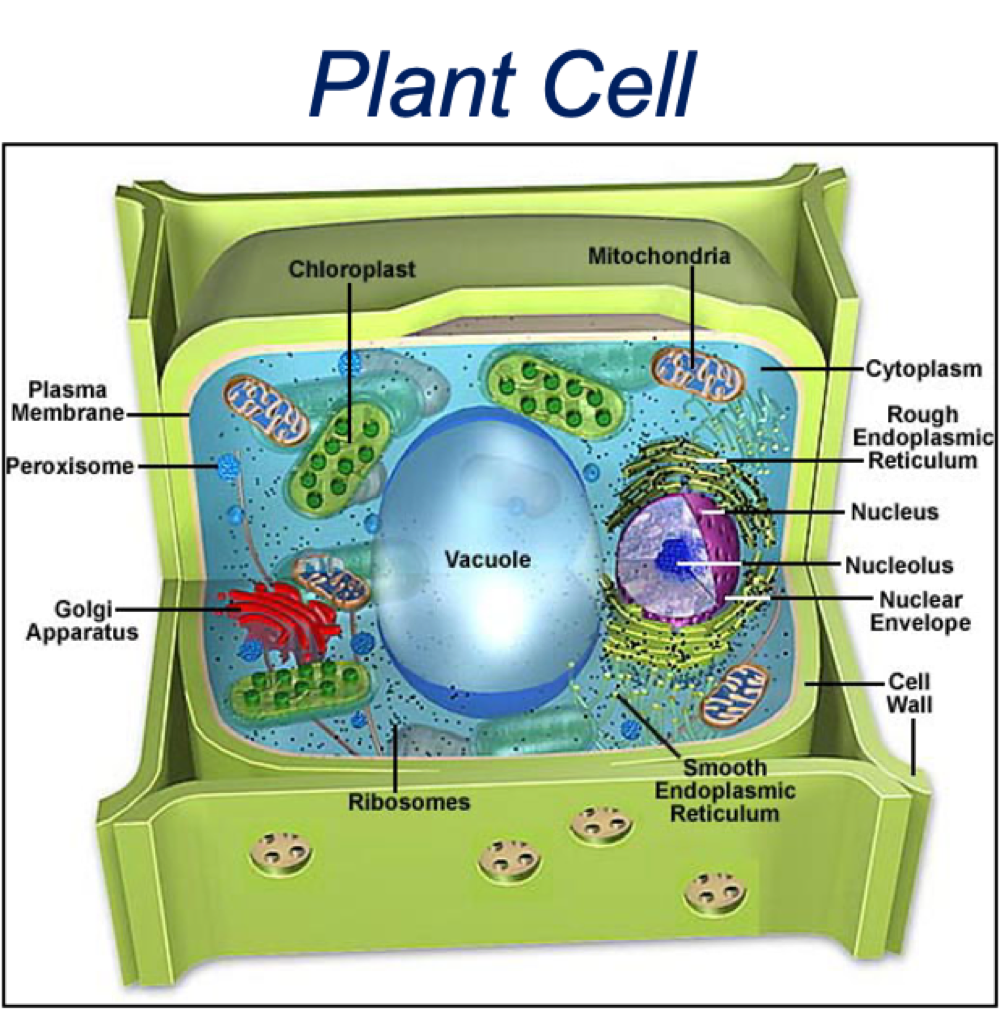
What are the differences between plant + animal cells?
plant cells larger than animals
additional structures in plants - chloroplast, cell wall, vacuole
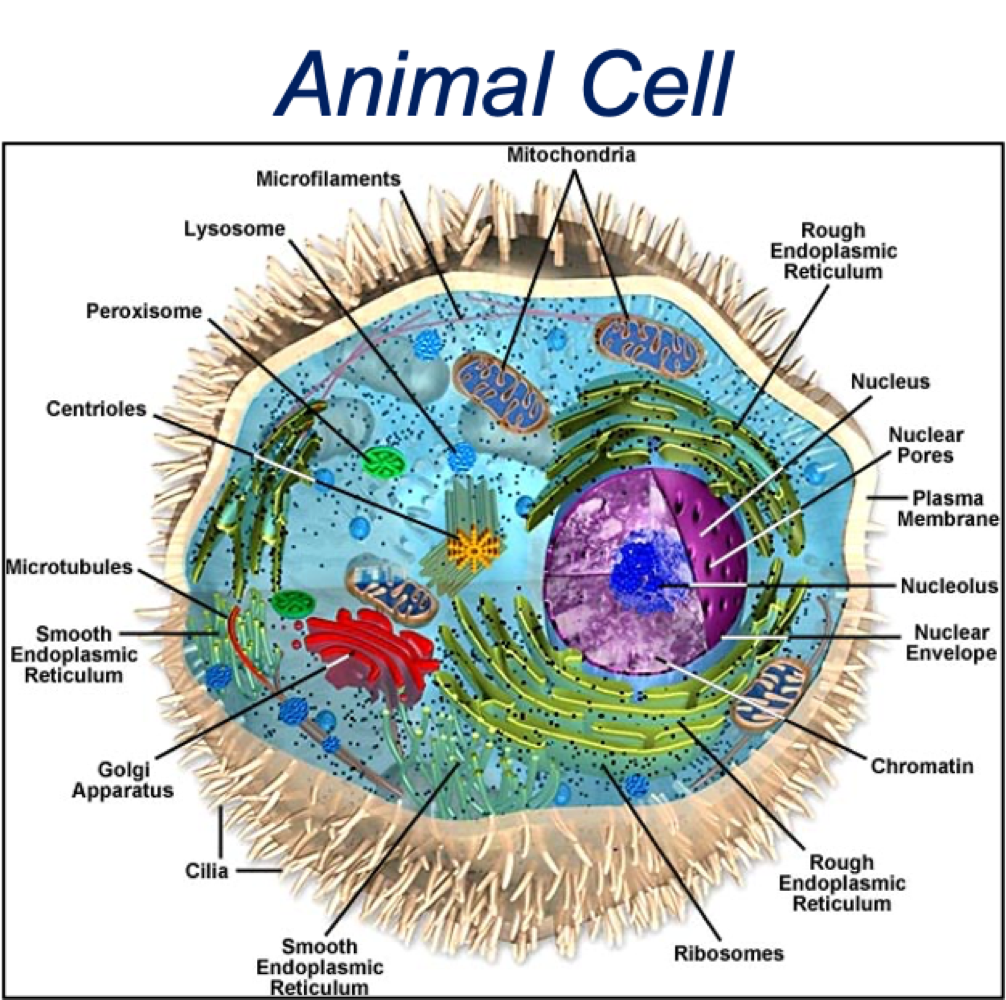
What are the double membraned organelles?
Nucleus, Mitochondria + chloroplast (M + C = own DNA)
What is the endosymbiosis theory?
Archaea genome similar to eukaryotic genome. Mitochondria + Chloroplast originate from the engulfment of bacteria by ancient archaea
What are the characteristics of the nucleus?
contains genetic information - DNA
DNA organised into chromosomes
Nuclear envelope = double membrane
What is the structure of the nucleus?
nuclear envelope = two membranes
outer nuclear envelope
perinuclear space
inner nuclear envelope
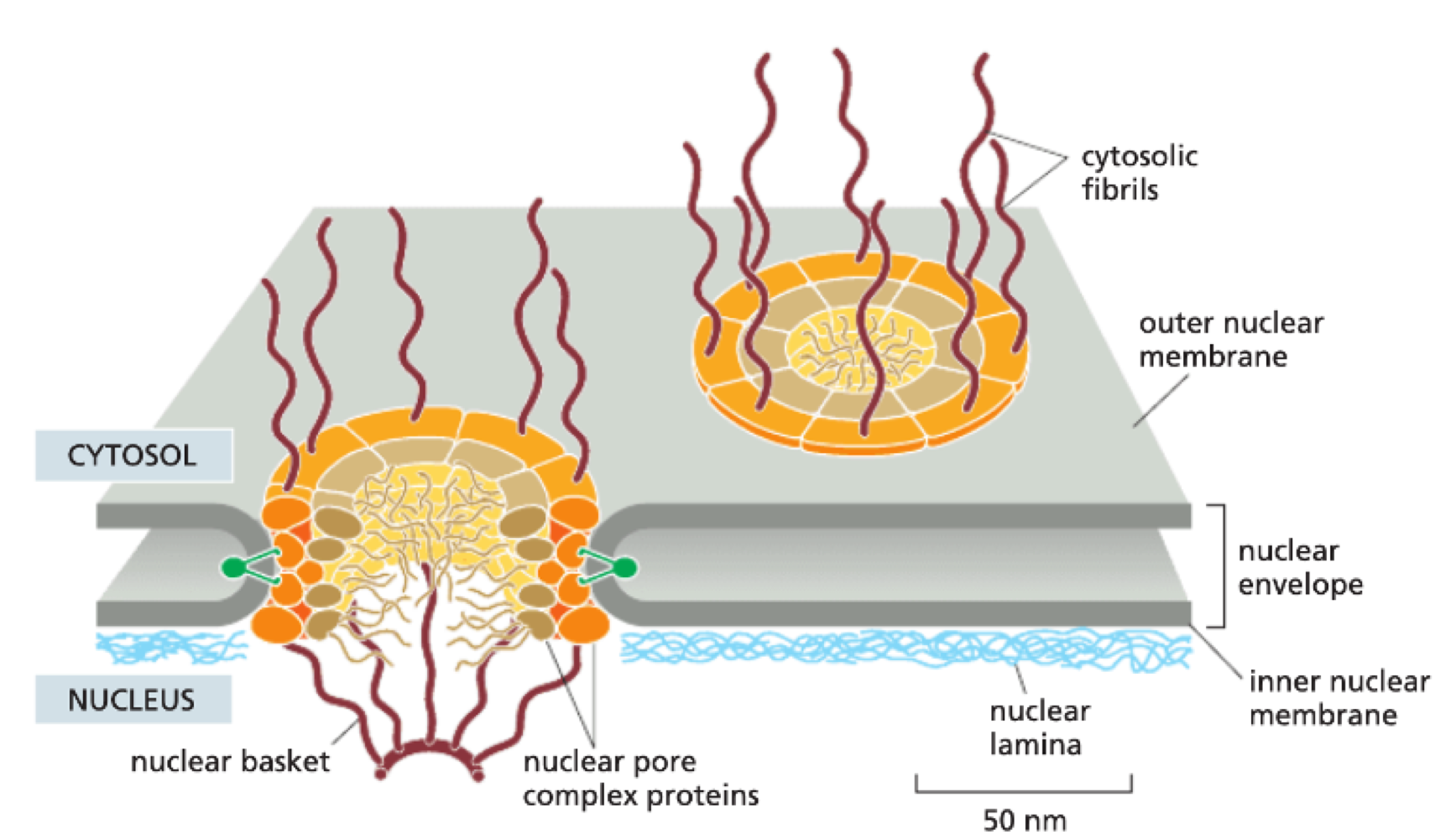
What are nuclear pores?
point of fusion between inner + outer nuclear membrane
gated by Nuclear Pore Complex (NPC) - made of 30 different proteins
How does the nuclear pore control what can enter/exit nucleus?
small molecules can diffuse by passive diffusion - large molecules cant enter passively
proteins created in cytosol can be transported through NPC if they have a Nuclear Localisation Signal (specific amino acid sequence)
Importin = recognises NLS
Binding of Importin to NLS allows transport through NPC
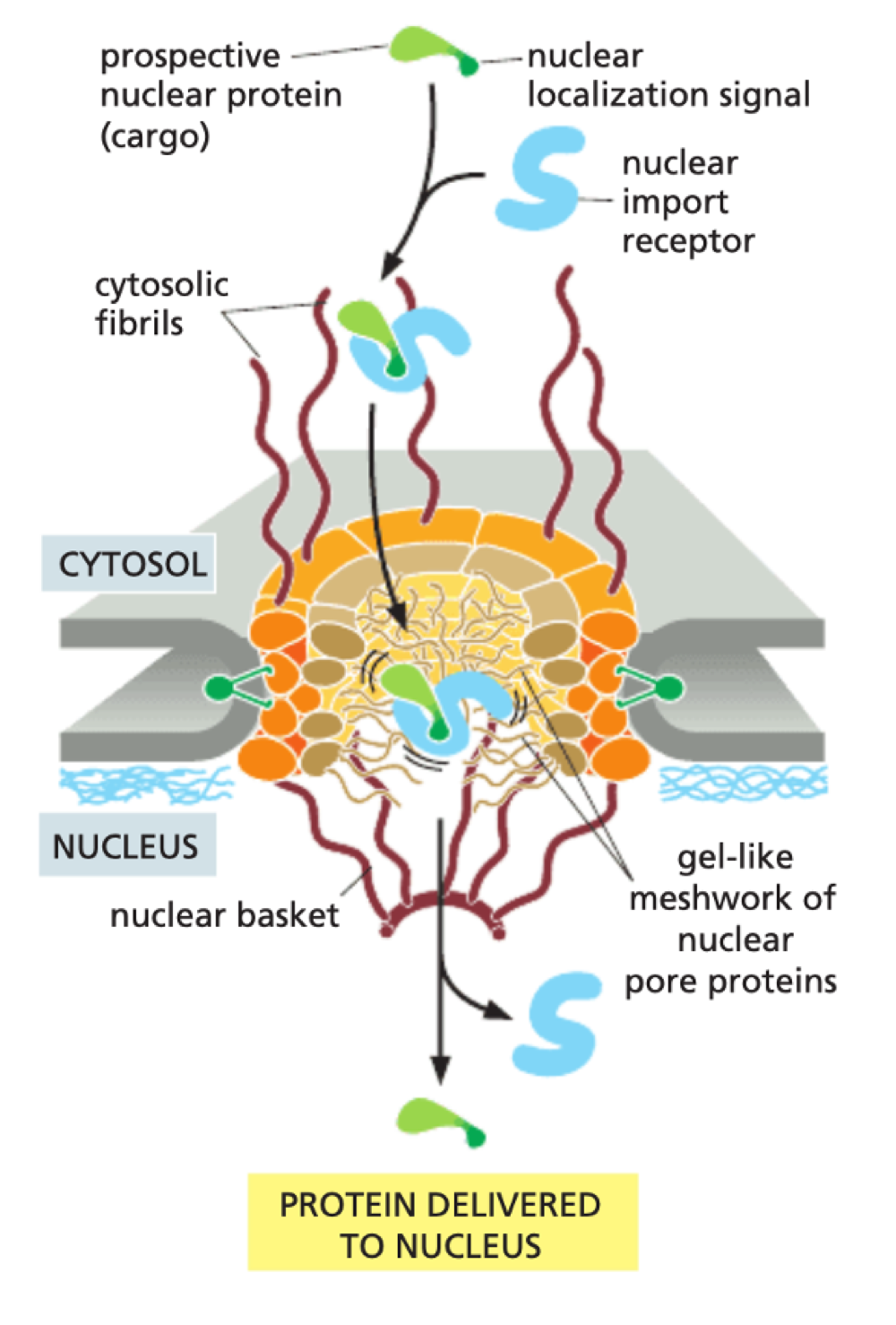
How does nuclear import use Ran-GTPase?
In cytosol - release of Ran-GDP allows protein to bind to importin
In nucleus - Ran-GTP binds to receptor to release protein
GTP hydrolysed to GDP as returns to cytosol
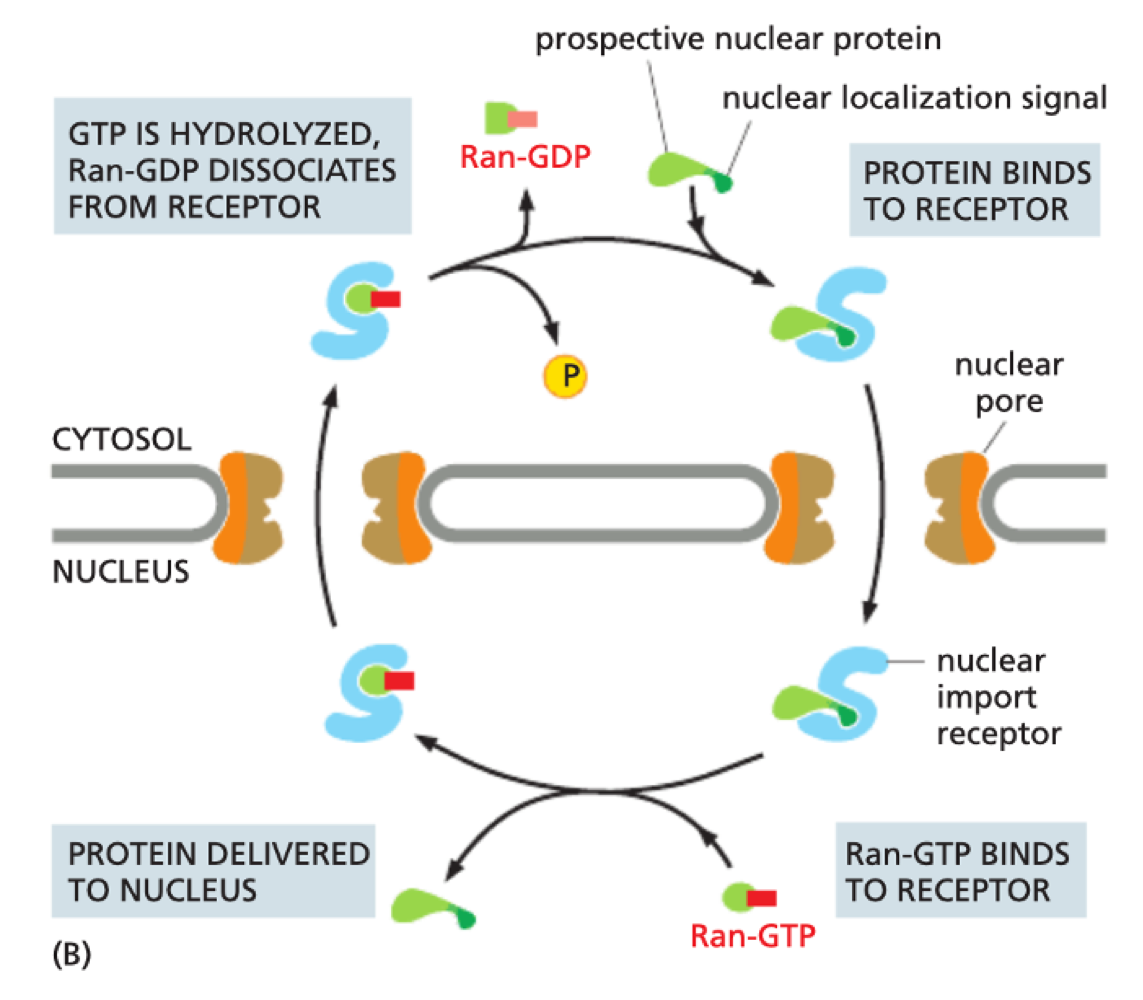
What is the process of nuclear export?
protein being exported has a Nuclear - Export Sequence (NES)
NES binds to Exportin
also requires Ran-GTPase + Ran-GTP/Ran-GDP gradient across nuclear membrane
important for export of RNA
What are the characteristics of the the nuclear lamina?
tough fibrous mesh under inner membrane
made of intermediate filament
acts as scaffold to maintain shape of nucleus
disassembles + reforms during division
What is the structure of chromosomes?
Chromatins (DNA wrapped in proteins - histones) associates with nuclear lamina. Each chromatin has its own discrete location - not randomly distributed
What is the nucleolus?
It is a biomolecular condensate - liquid like behaviour. Centre for ribosomal RNA (rRNA) production. Responsible for synthesis + assembly of RNA and protein to for ribosome.
What is the structure of endoplasmic reticulum?
Continuous structure with outer nuclear membrane
interconnected tubes + flattened sacs = high SA
share single internal space - ER lumen
10% of cell volume
What are the type of endoplasmic reticulum?
Rough ER - ribosomes on surface = protein synthesis
Smooth ER = biosynthesis + metabolism of lipids
Transitional ER = producing vesicles with protein or lipid for transport to Golgi
How does rough ER carry out its function?
proteins are translocated into ER lumen while being translated from mRNA. Carries out simple modification of proteins + folding quality check.
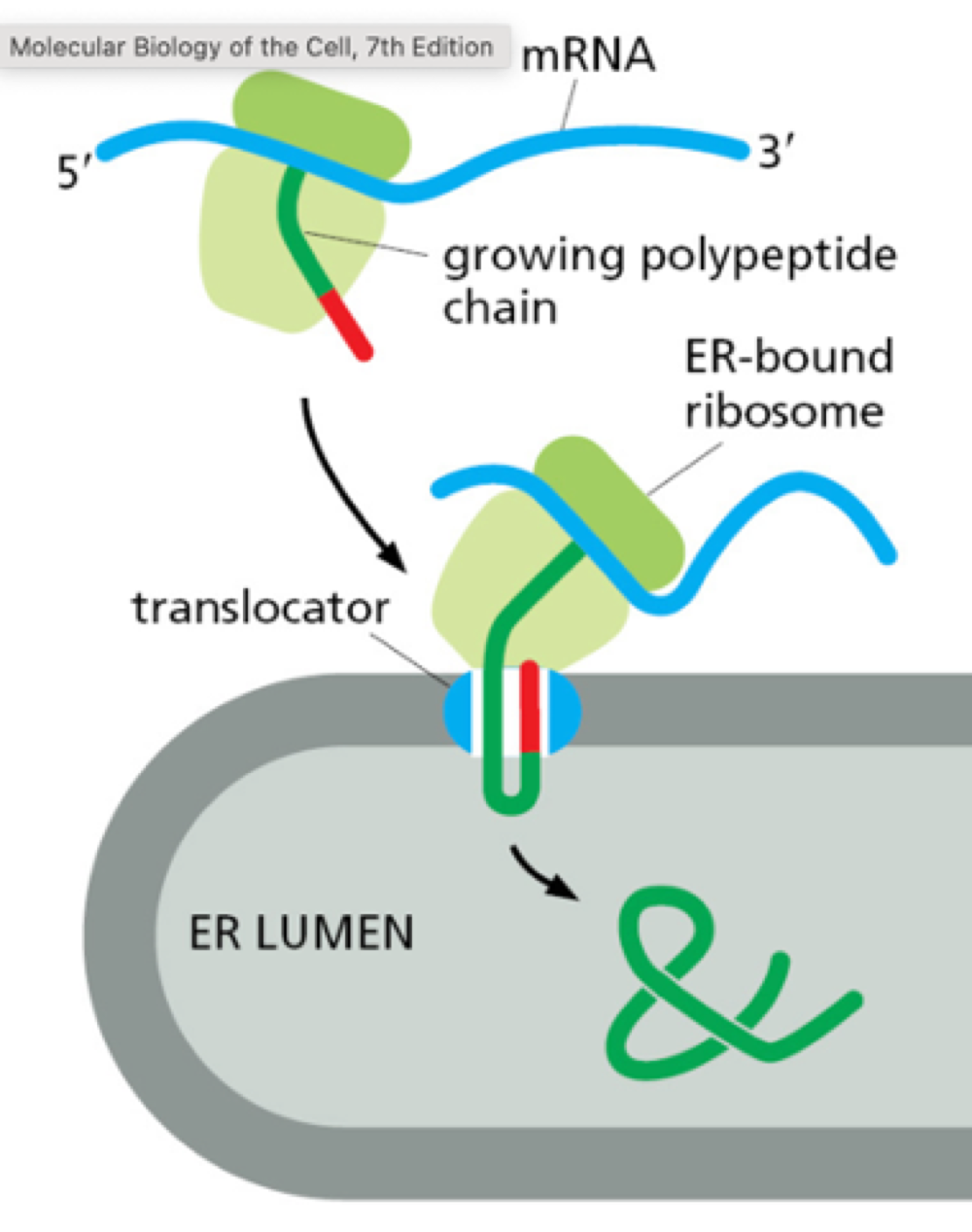
Where is smooth ER found?
found in cells involved in lipid metabolism. Synthesises lipids for production of lipoprotein also contains enzyme for drug detoxification.
Muscles - special type = sarcoplasmic reticulum
stores calcium ions to regulate muscle contraction
What is the structure of the golgi apparatus?
collection of flattened, membrane enclosed sac = cisternae
Cis complex = adjacent to ER + recieves proteins from ER
Medial cisternae = in middle
Trans complex = towards plasma membrane
What are the two models that explain vesicle trafficking through cell?
Vesicular transport model:
resident enzyme stays in same cistern (proteins that modify proteins)
cargo proteins are moved between each cisternae
Cisternae maturation model:
cargo proteins stay in same cisternae
resident proteins move between cisternae - cisternae mature between through layer
Why are transport vesicles coated?
determines where they go
Clathrin coated - from Golgi, end-some, plasma membrane
COPI coated - from Golgi
COPII coated - from ER
Retromer coated - endosome retrieval to Golgi
How do proteins move across membrane barrier?
Gated transport - e.g. nuclear pore complex
Protein translocation - proteins are synthesised into compartments
Vesicular transport - membrane-bound vesicles ferry proteins from one compartment to another
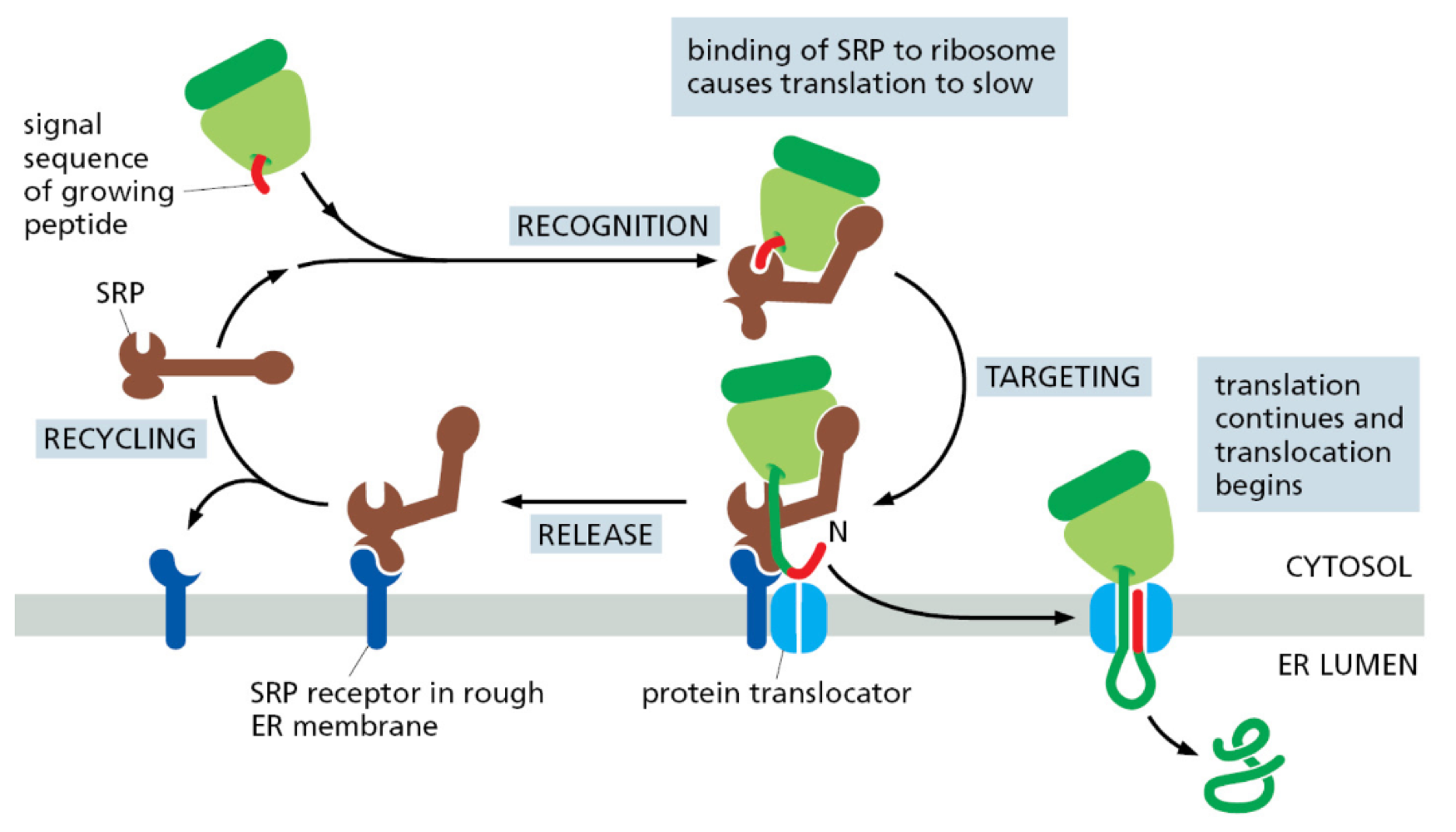
How does co-translation translocation occur?
newly made peptide with ER signal sequence bind to signal recognition particle (SRP)
SRP guides ribosome + RNA to translocator by binding to SRP receptor on plasma membrane
protein translation occurs through translocator into ER lumen
Proteins unfold during translocation and refold after
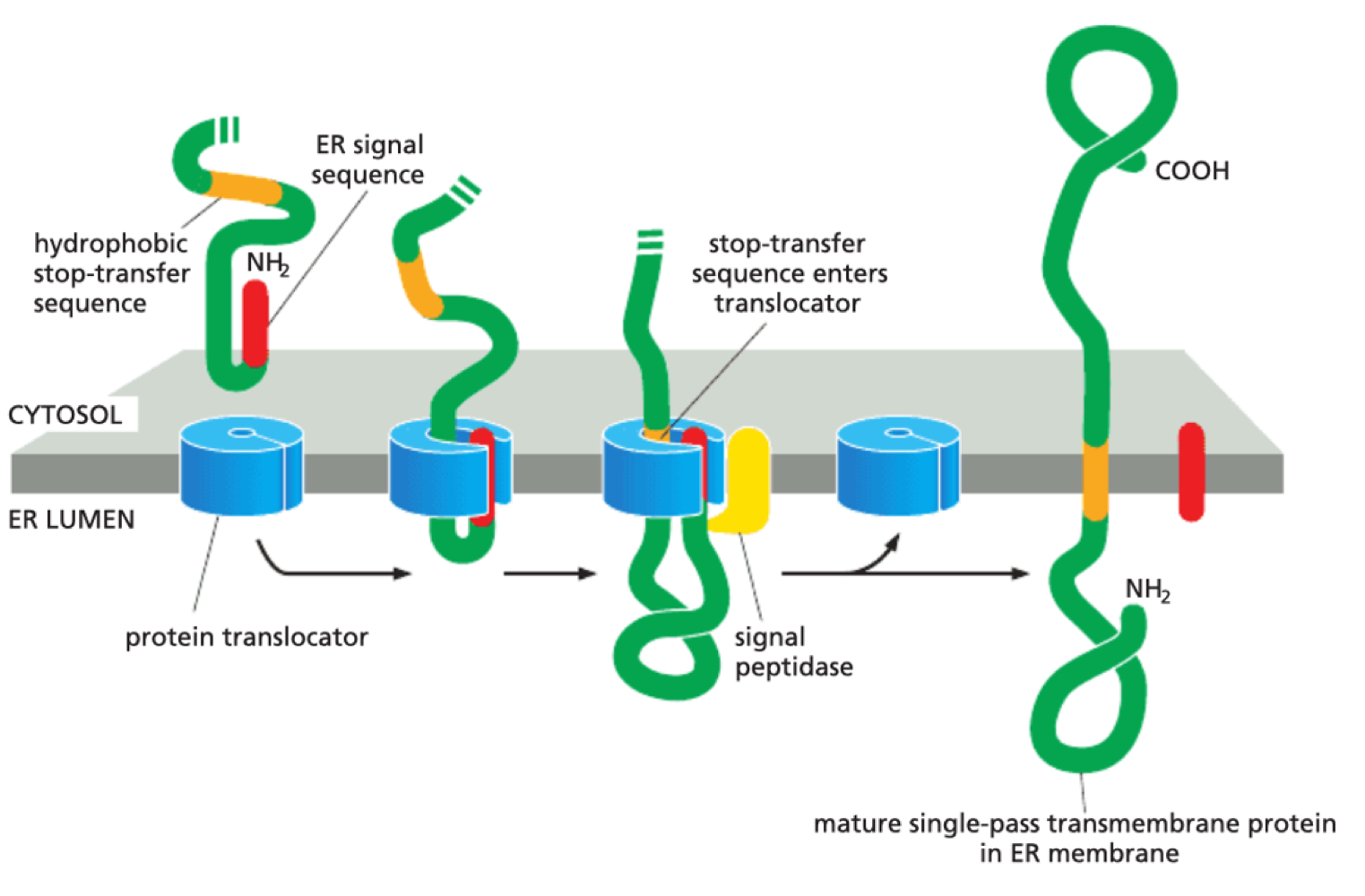
How are transmembrane proteins produced?
Stop transfer sequence prevents complete entry into ER lumen - proteins become transmembrane
How are vesicles coated?
cargo selection
memory bending
protein coating
coat disassembly
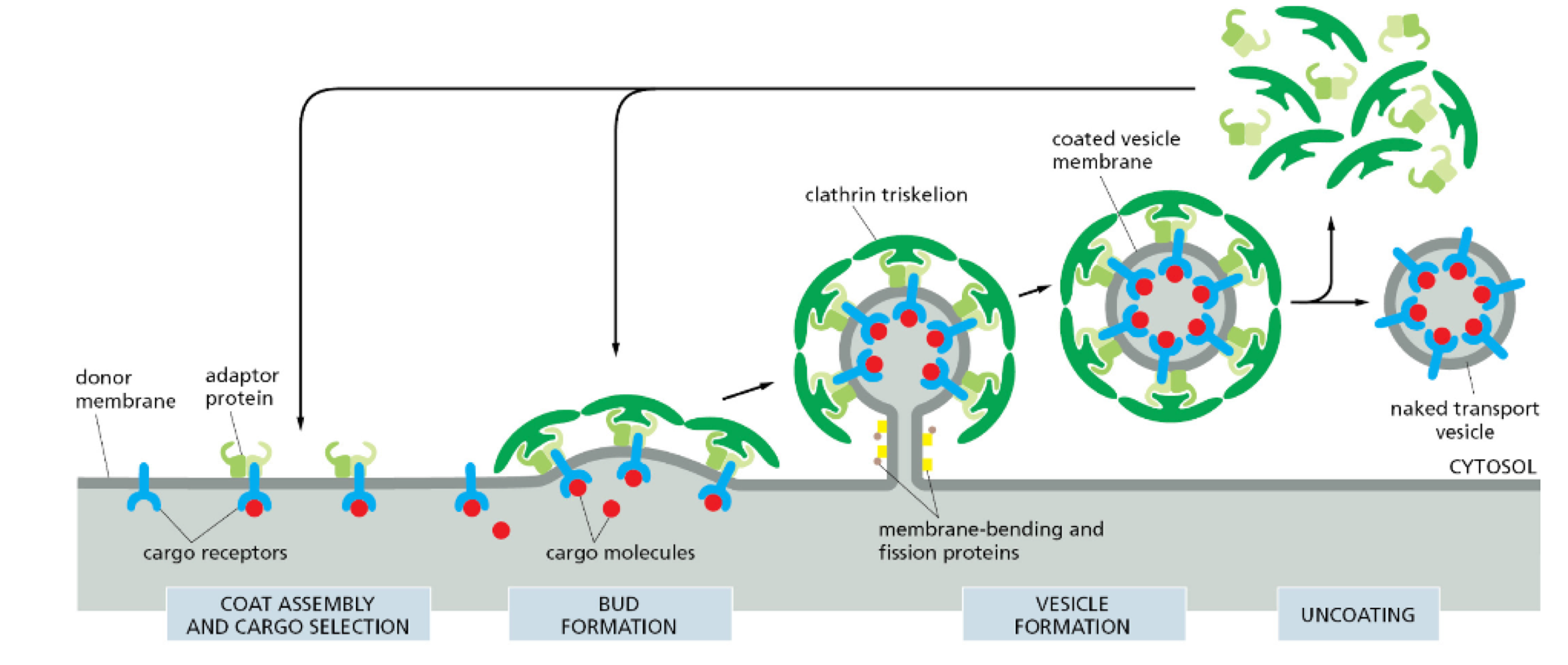
How do vesicles know where to go?
Vesicle coat = address label
Phospholipid profile = post code (differently phosphorylated)
Rab GTPases = postman
guides vesicle to destination
different ones guide to different locations
GTP-binding = active
GDP-binding = inactive
How does vesicle-membrane fusion take place?
Rab GTPase interacts with tethering proteins on target membrane
v-snare + t-snare (transmembrane proteins) interacts - highly complementary + specific
v + t-snare ‘zipping’ is what forces membranes together by removing water between two phospholipid bilayers
v + t-snare determine if vesicle fuses in correct destination
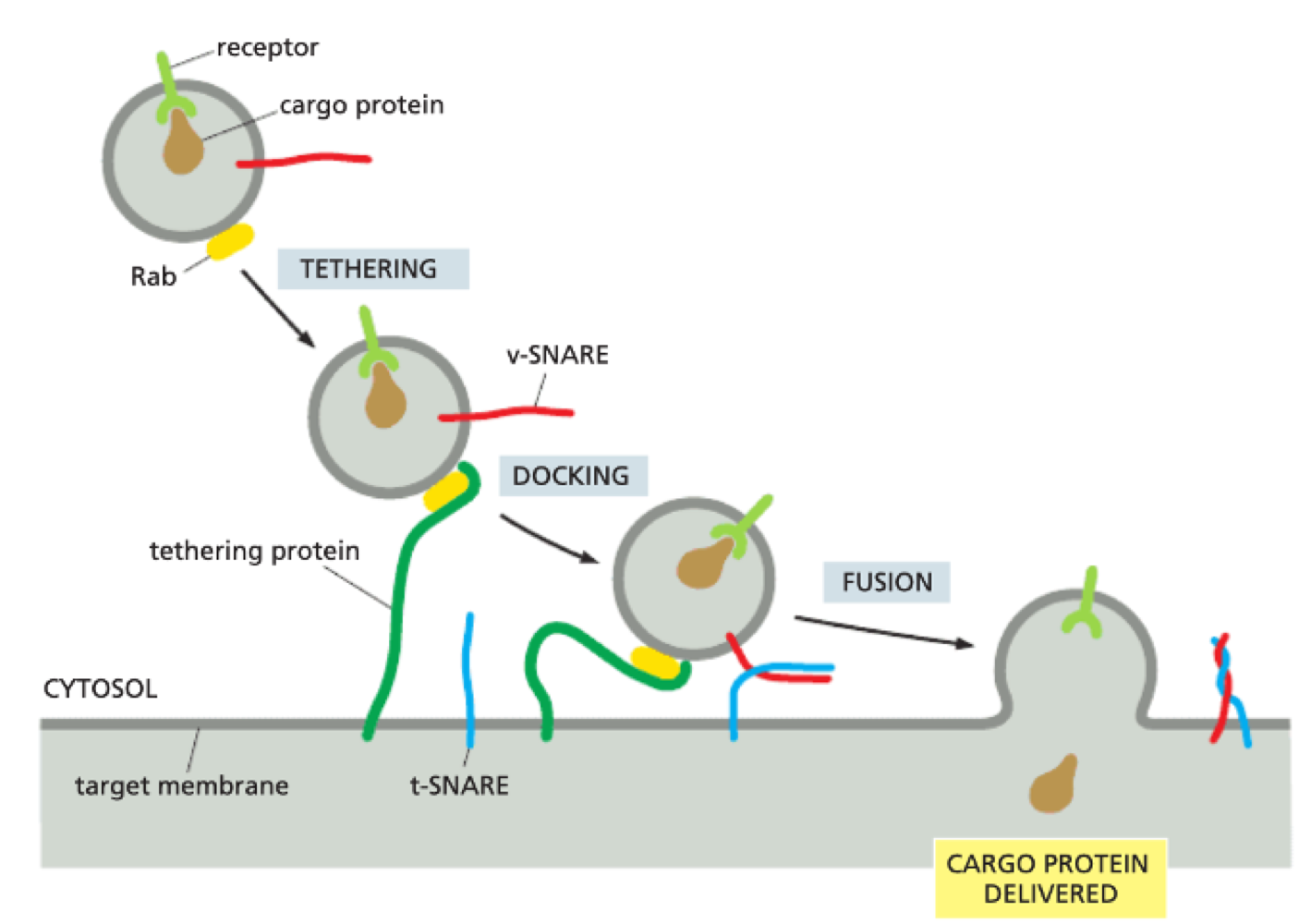
What is exocytosis?
movement out of a compartment
What is endocytosis?
movement to a compartment
What is the process of exocytosis?
delivery of newly made proteins, lipids + carbohydrates to cell surface
ER → Golgi → Plasma membrane
What are the types of exocytosis?
Constitutive exocytosis - continuous supply of plasma membrane with new lipid + protein
Regulated exocytosis - secretary vesicles stored + released upon stimulus
How does regulated exocytosis occur in insulin secretion?
insulin produced + stored in vesicles in pancreatic beta-cells
glucose uptake results in increased ATP
ATP inhibits potassium channels causing membrane depolarisation
opening up calcium channels that trigger exocytosis of insulin-containing vesicles.
What is the process of endocytosis?
internalisation of large/small molecules and liquid
phagocytosis = large
pinocytosis = small
endocytic vesicles bud inwards and is delivered to either lysosome or endosome.
lysosome = digestion, endosome = recycled to plasma membrane
What is the process of phagocytosis?
Internalisation of large particles e.g. bacteria. - lysosomes
Immune cells that carry out phagocytosis = macrophages + neutrophils
cell surface receptors on phagocytic cell must be activated before phagocytosis
important for scavenging dead/damaged cells
What is the process of pinocytosis?
uptake of small volume of extracellular fluid + bits of own plasma membrane
cell volume + total surface area dont change
How do viruses enter cells by endocytosis?
virus can hack receptor-mediated endocytosis trafficking to enter host cell. Virus use receptor-binding domain of spike protein on its surface to bind to receptor on plasma membrane and activate endocytosis.
What are the characteristics of endosomes?
intracellular sorting organelles
connected membrane tubes + large vesicles
early endosome = under plasma membrane
late endosome = closer to nucleus
acidic interior maintained by ATP driven H+ pump
role:
recycling membrane receptors
lysosomal degradation
What are the characteristics of lysosomes?
degrade proteins, nucleic acids, lipids + oligosaccharides
contain hydrolytic enzymes - optimally active in acidic environment
H+ pump make lumen acidic
have specialised transporters to export useful metabolites out to cytosol - recycling
What are the functions of the plant vacuole?
storage - nutrients + waste products
degredation compartment
cell size increase
control turgor pressure
maintain pH homeostasis
What are the characteristics of peroxisomes?
single membrane enclosed organelles - very small
contain oxidative enzymes: catalase, urate oxidase
produces hydrogen peroxide
catalases use hydrogen peroxide to oxidise lipids + detoxify harmful metabolites
myelin sheath = highly dependant on phospholipid metabolism
What are the characteristics of mitochondria?
highly dynamic
highly plastic
vary among cells - rbc = none, liver cells = more than 1000
often associated with cytoskeleton
What is the double membrane structure of mitochondria?
outer membrane
contains large, channel proteins e.g. porin
permeable to small molecules
Inner membrane
folded into cristae
contains proteins for electron-transport chain and ATP synthesis
contain transport proteins into + out of mitochondria matrix

What is the structure of the mitochondial matrix?
highly concentrated mix of enzymes + dna
mitochondrial DNA genome
mitochondrial ribosomes + tRNA
enzymes for oxidation of pyruvate + fatty acids + for citric acid cycle
What is the structure of the intermembrane space?
concentration of small molecules - same as cytosol
higher concentration of proteins - cytochrome C
release of cytochrome C - apoptosis
What makes mitochondria dynamic?
they can divide by fission + fusion, and move by cytoskeleton
How is ATP produced in mitochondria?
occurs in mitochonrial inner membrane - OXIDATIVE PHOSPHORYLATION
Acetyl CoA production
Activated electron carrier: NADH + FADH2
stepwise electron movement along electron transport chain
electrochemical gradient
proton flow back into mitochondrial matrix
‘turn’ ATP synthase + create ATP
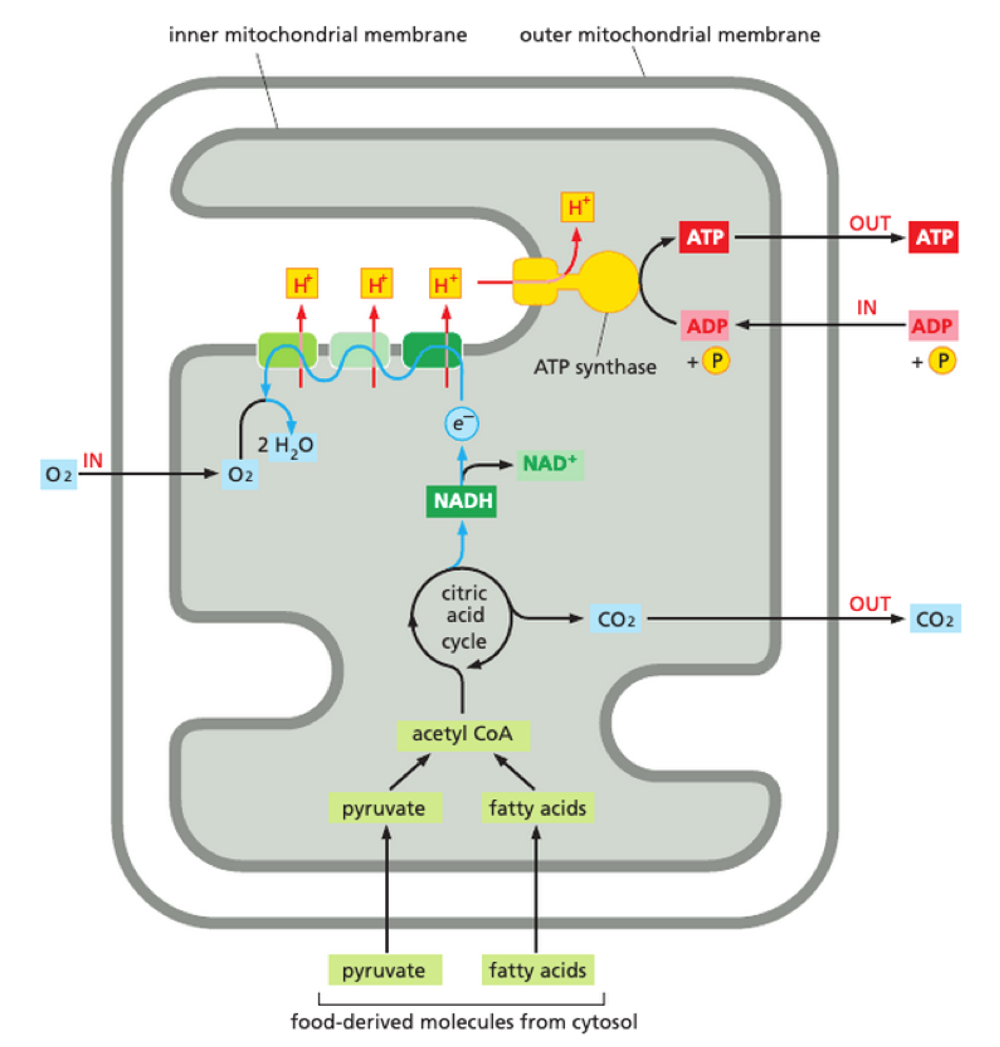
What is the electron transport chain?
4 large protein complexes - 3 = proton pumps
during electron transfer from NADH, a hydride ion is removed and converted to a proton + 2 electrons
electrons donated to electron transport chain
electron moves through complex - induces conformational changes in protein = pumps proton from matrix to intermembrane space
energy to pump H+ from energy of electron transfer
build up of H+ in intermembrane space - creates electrochemical gradient
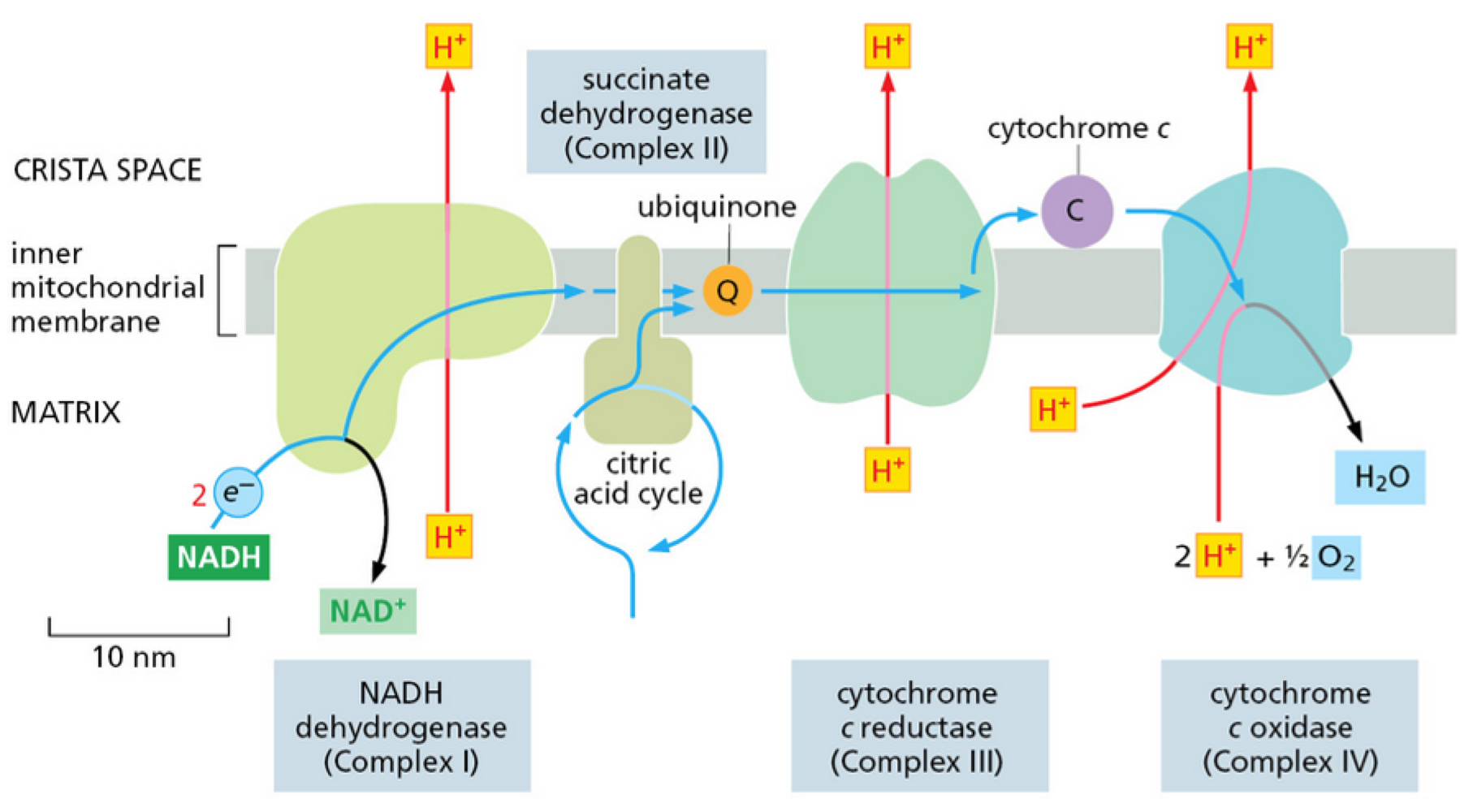
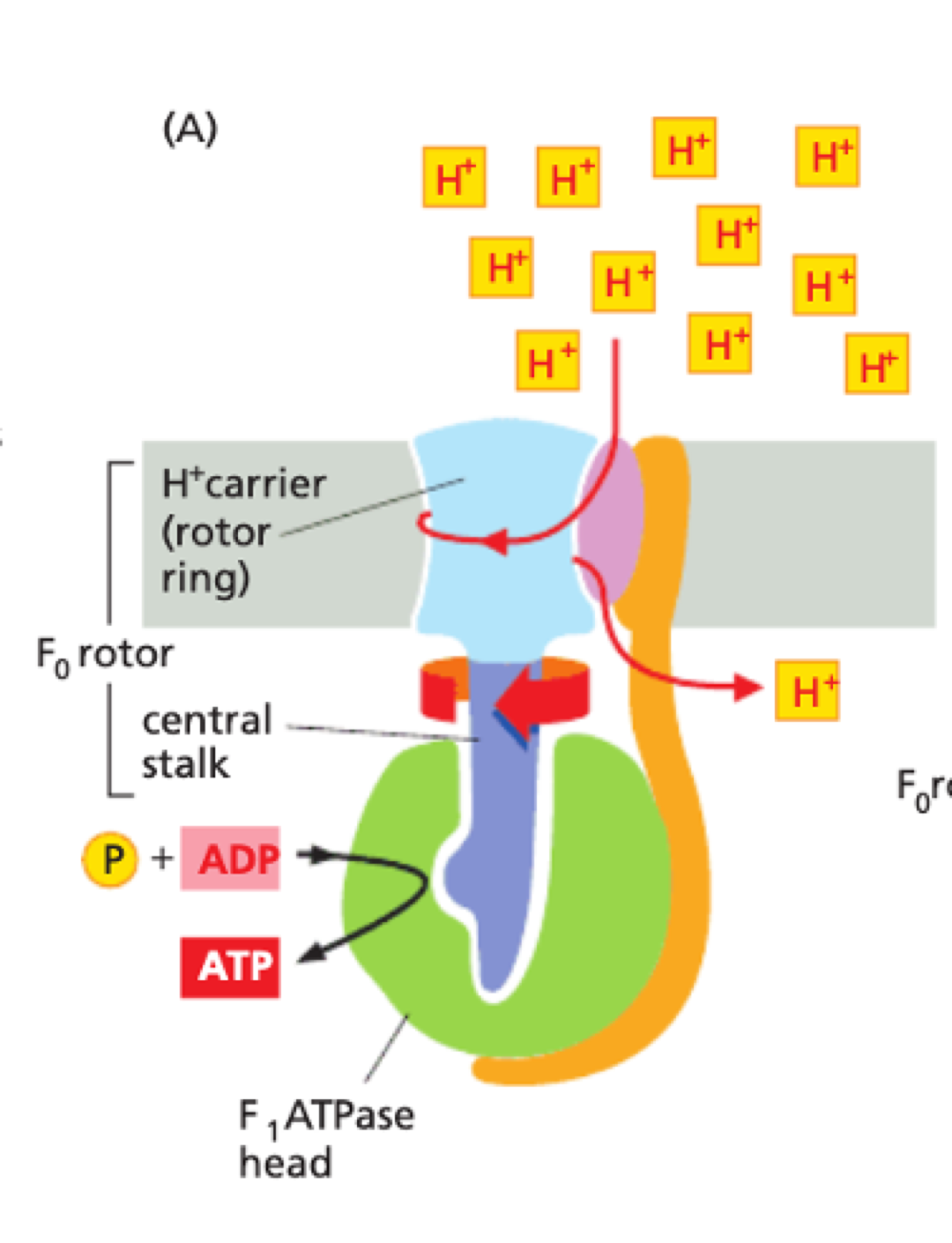
How does proton motive force make ATP?
membrane potential + pH gradient = proton motive force
H+ carrier spins rapidly within stationary head of F1 ATPase
F1 ATPase converts ADP to ATP
ATP synthesised in matrix is pumped back into intermembrane space
ADP back into matrix
done by a ADP/ATP carrier protein on inner membrane
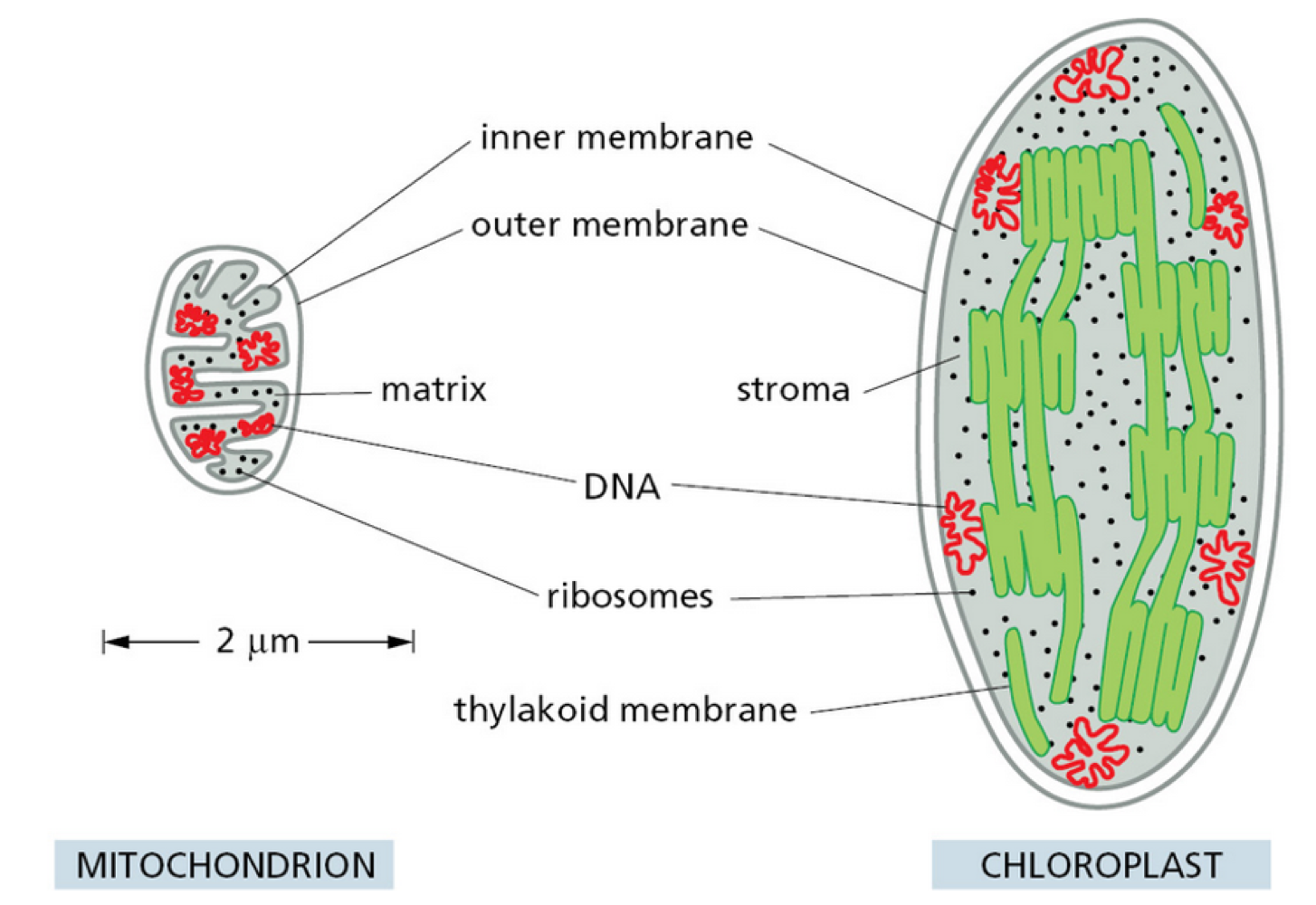
What are the similarities/differences between mitochondria + chloroplast?
chloroplast generally larger than mitochondria
similar structures:
double membrane
highly permeable outer membrane
tight intermembrane space
chloroplast have extra structure = thylakoid
What is the internal structure of chloroplast?
stacks of thylakoid = grana - contain chlorophyll
What are the characteristics of chlorophyll?
absorb red + green light
green light absorbed poorly - reflected
this is why plants look green
different chlorphylls have different absorption preferences
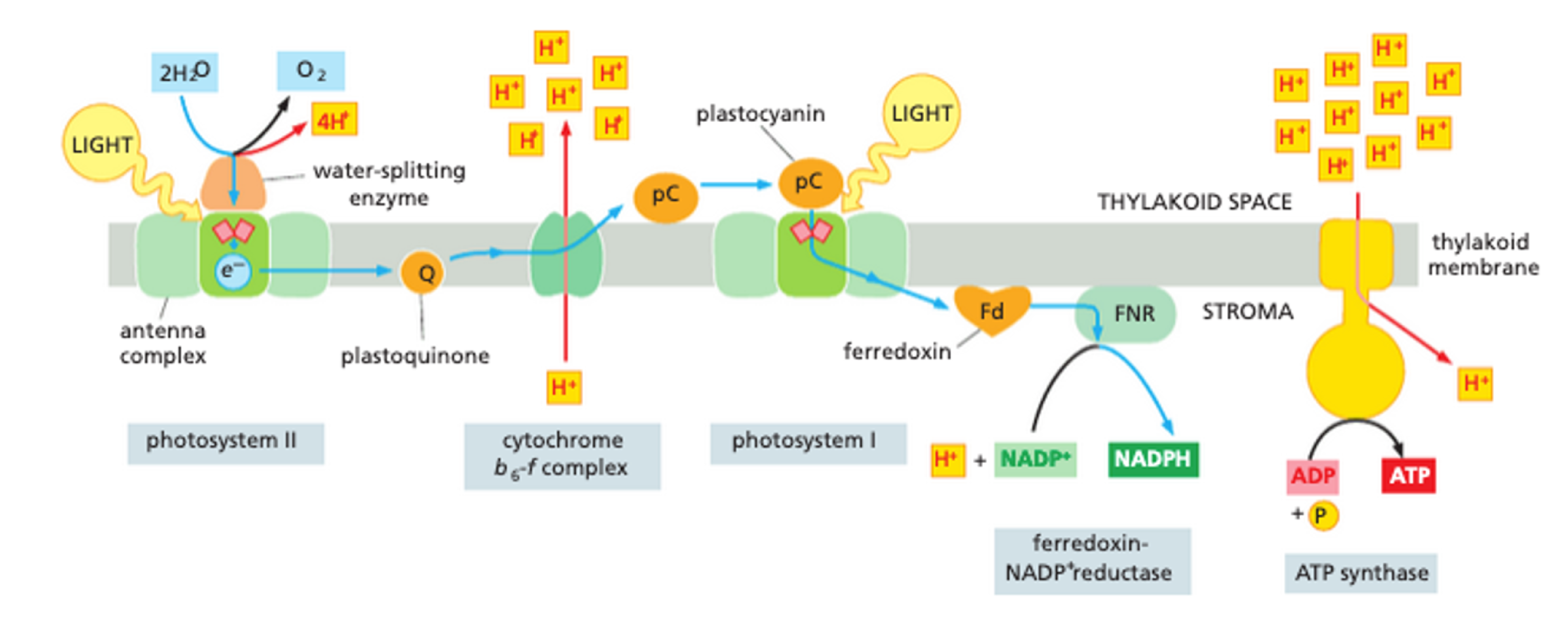
What is stage 1 of photosynthesis?
photosystem II = water splitting enzyme produces O2
excided electrons are passed down via electron carriers to photosystem I
electron proton gradient allows ATP synthesis
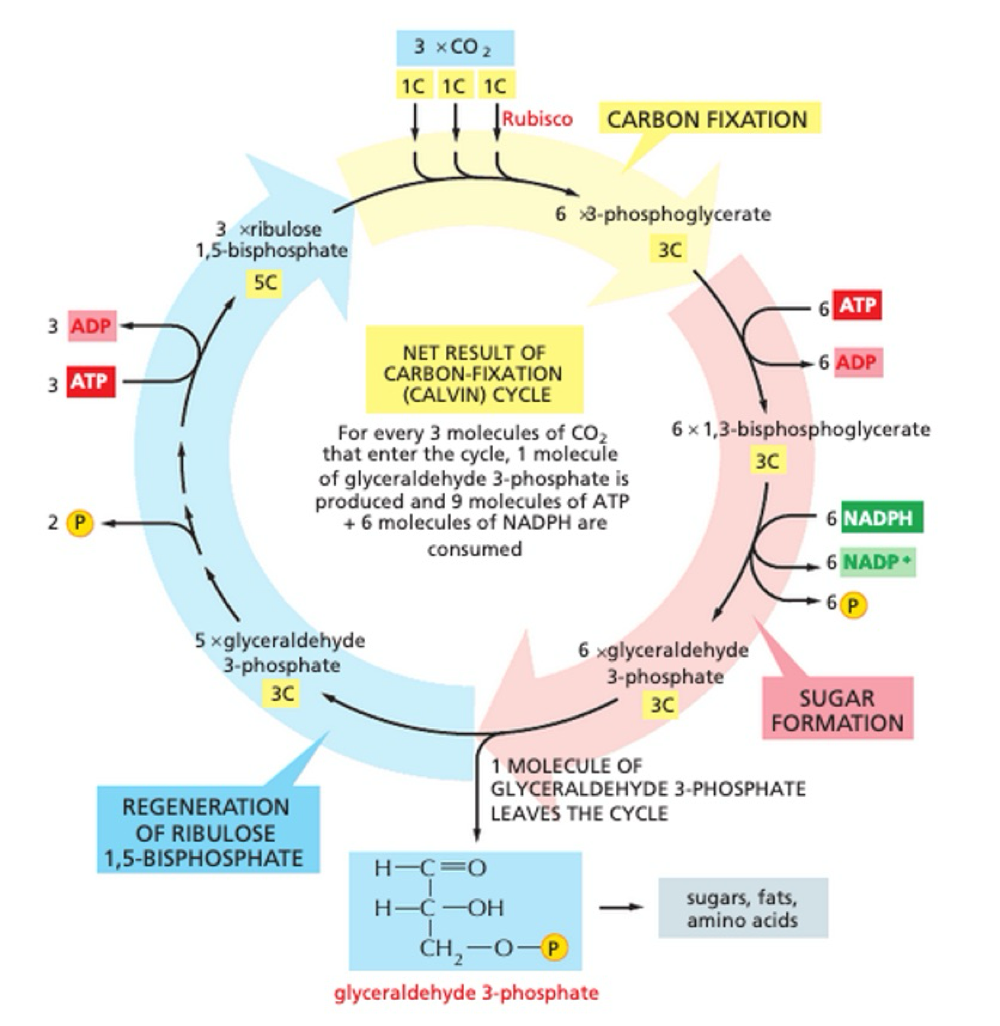
What is stage 2 of photosynthesis?
light independant reaction
thylakoid membrane is impermeable to ATP + NADPH - no diffusion back into thylakoid membrane
ATP and NADPH + CO2
used to produce simple 3-carbon sugar
exported + used as precursor for synthesis of other metabolites
What is the key enzyme for the light independent reaction?
Rubisco
slow + abundant
How do mitochondria + chloroplast collaborate?
sugars made in chloroplast can be stored as starch or exported
exported can be broken down + imported into mitochondria for efficient ATP synthesis
O2 released from chloroplast - used in oxidative phosphorylation
CO2 released from mitochondria - used in carbon fixation
How are ribosomes structured?
ribozyme = RNA enzyme
two subunits - large + small
by mass: 2/3 rRNA + 1/3 ribosomal protein
eukaryotic 80s ribosomes - 40s subunit + 60s subunit
assembled in the nucleolus
How are ribosomes made?
rRNA is folded into highly compact + precise 3D structure
ribosomal protein on surface + fills gaps in folded RNA core
contains 4 binding sites:
1 for mRNA
3 for tRNA - only 2 occupied at once
How do ribosomes carry out translation?
tRNA ‘charged’ with amino acid form base pair with complementary codon on mRNA
A + P sites very close - no base between
peptidyl transferase activity
large subunit translocate relative to smal subunit
position tRNA into exit site
small subunit translocate exactly 3 bases back to original position
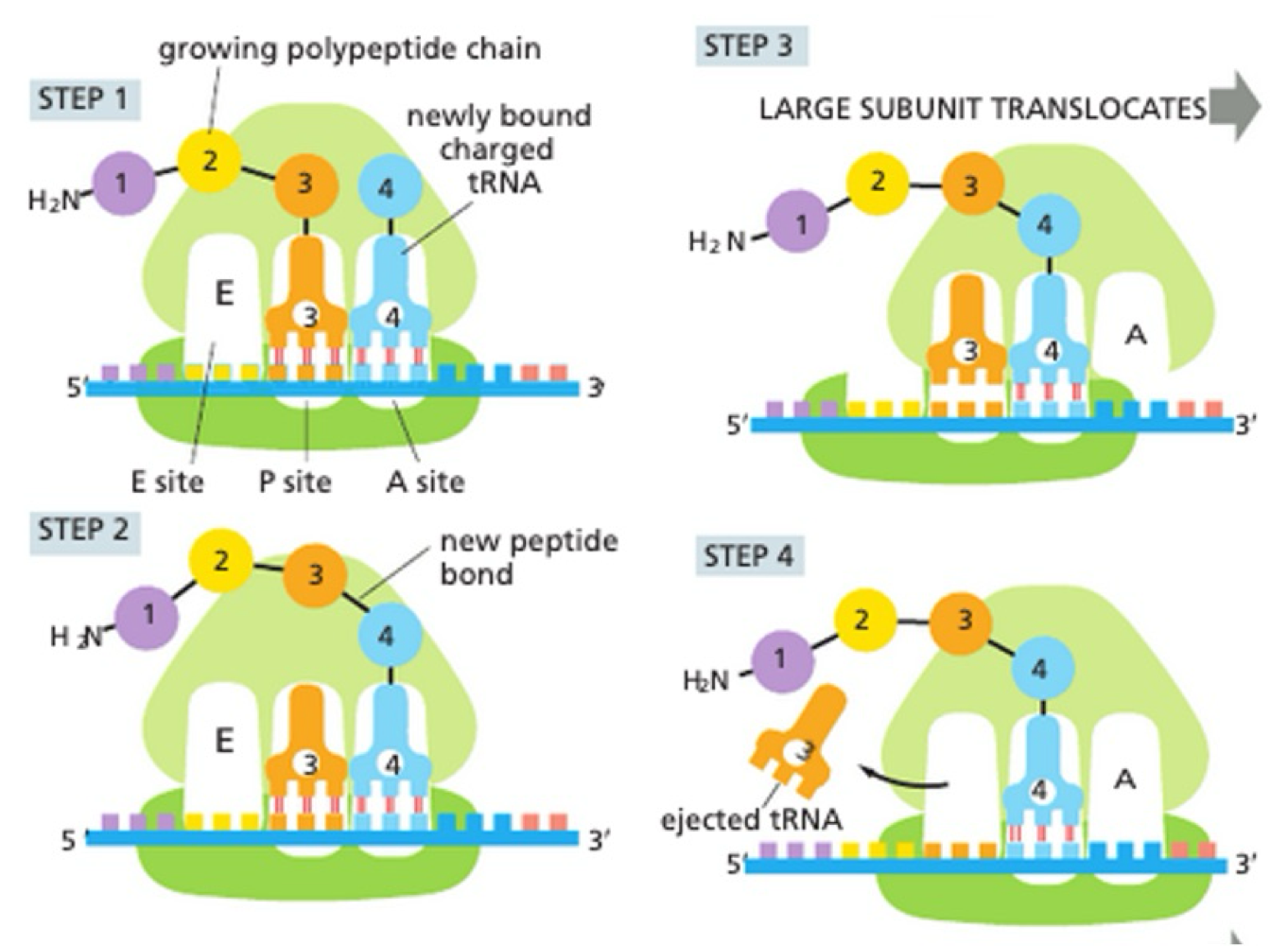
How does ribosome reset after translation?
ejection of used tRNA
empty A site ready for next ‘charged’ tRNA to bind
repeated until stop codon reached
What determines translation speed?
single mRNA can have multiple translations simultaneously - aslong as previous ribosome moves out the way, new one can bind
polyribosomes = especially important for highly-tanslated proteins
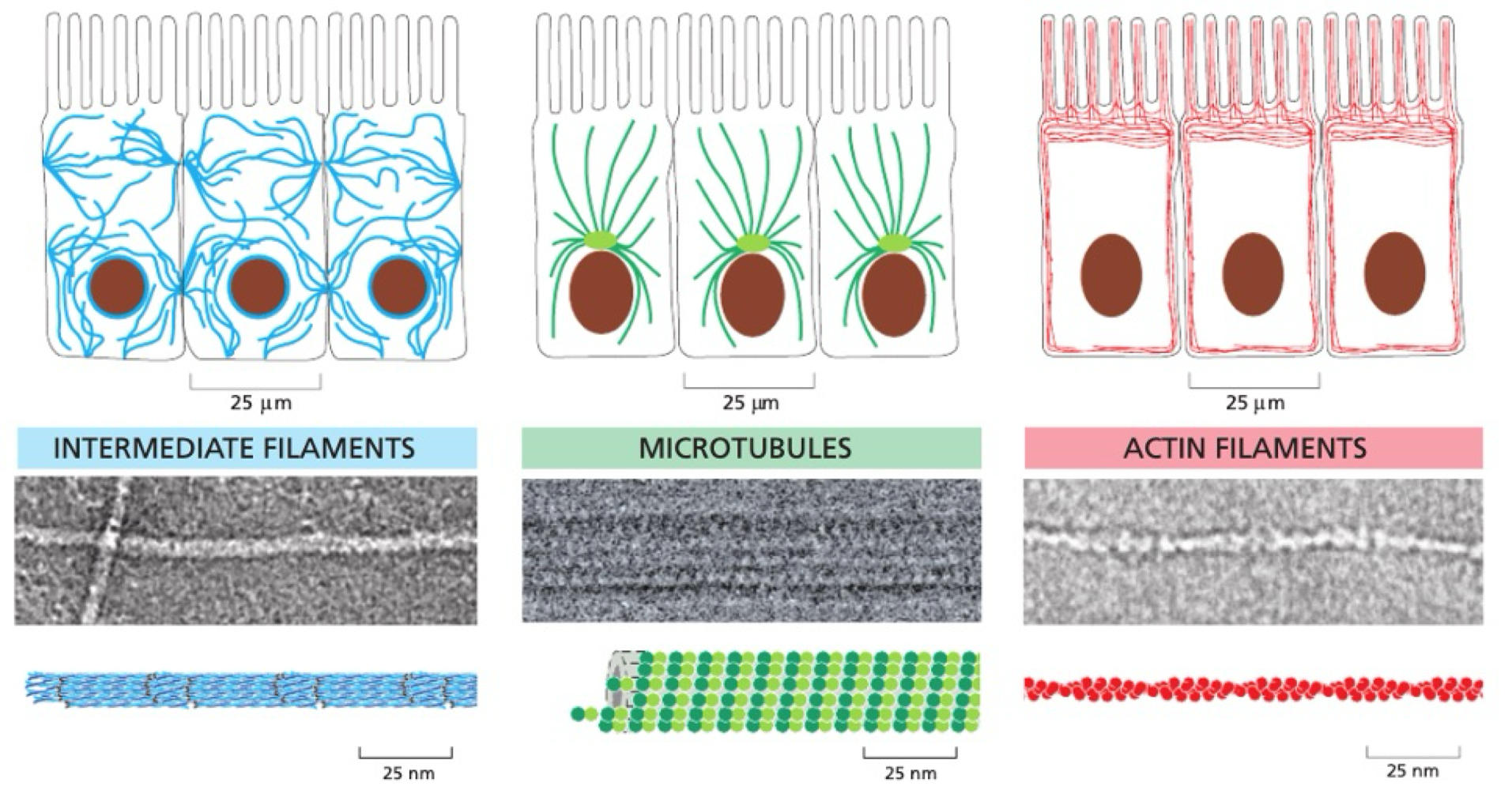
What are the types of cytoskeleton?
intermediate filaments, microtubules + actin filaments
dynamic
flexible
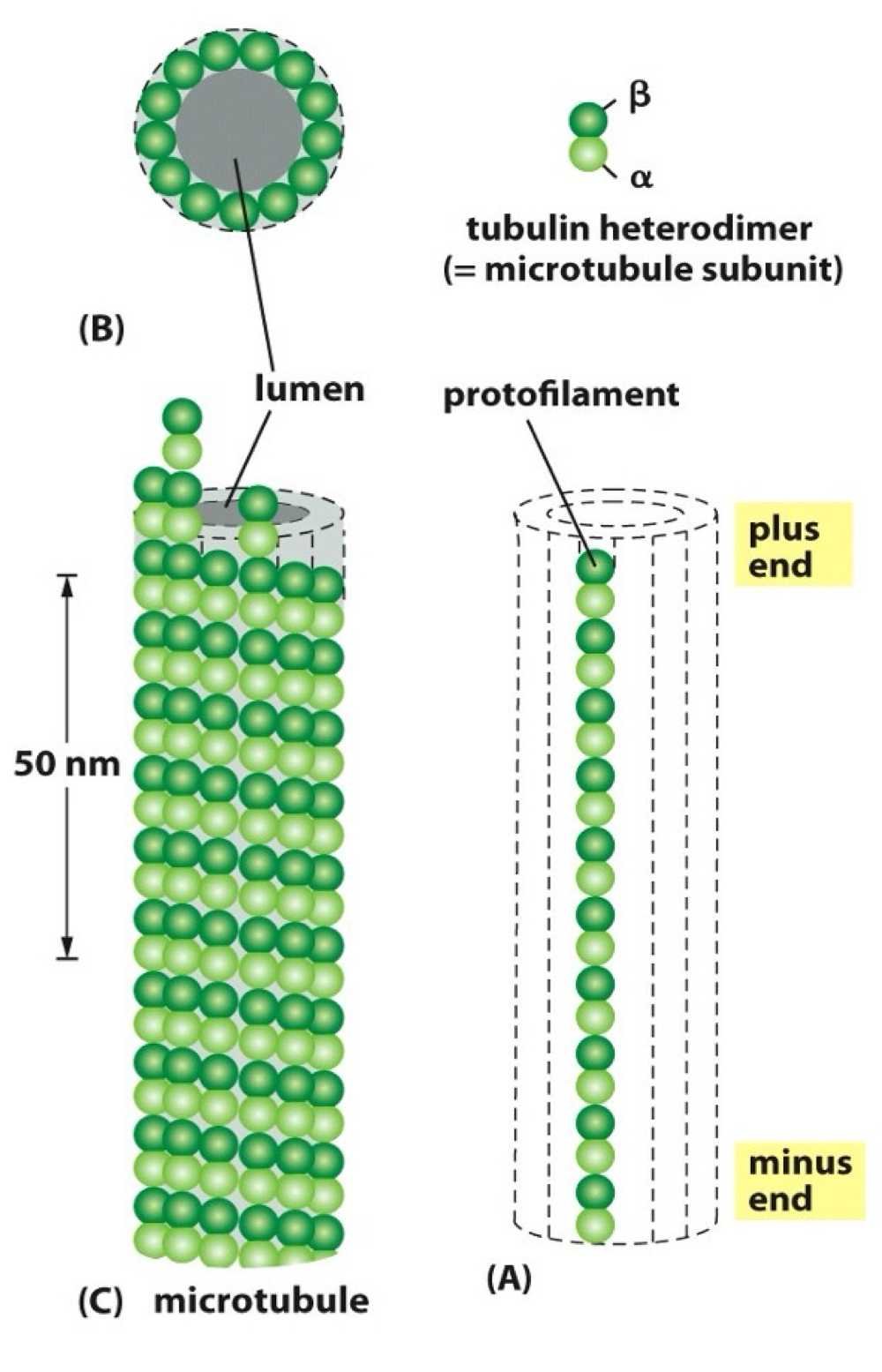
What is the structure of microtubules?
hollow tube
13 protofilaments per microtubule
made from α + β tubulin heterodimers
‘plus’ + ‘minus’ ends - β tubulin at + end
each dimer makes 2x lateral contacts + 2x longitudinal contacts
What are the characteristics of α + β tubulin?
highly conserved - found in all organisms in a similar structure
each bind to one GTP molecule
α tubulin bound GTP = not hydrolysed
β tubulin = hydrolysed to GDP
How do you describe microtubule dynamic?
Nucleation - initiation of MT = start from scratch
Polymerisation - growth of MT = elongation of existing
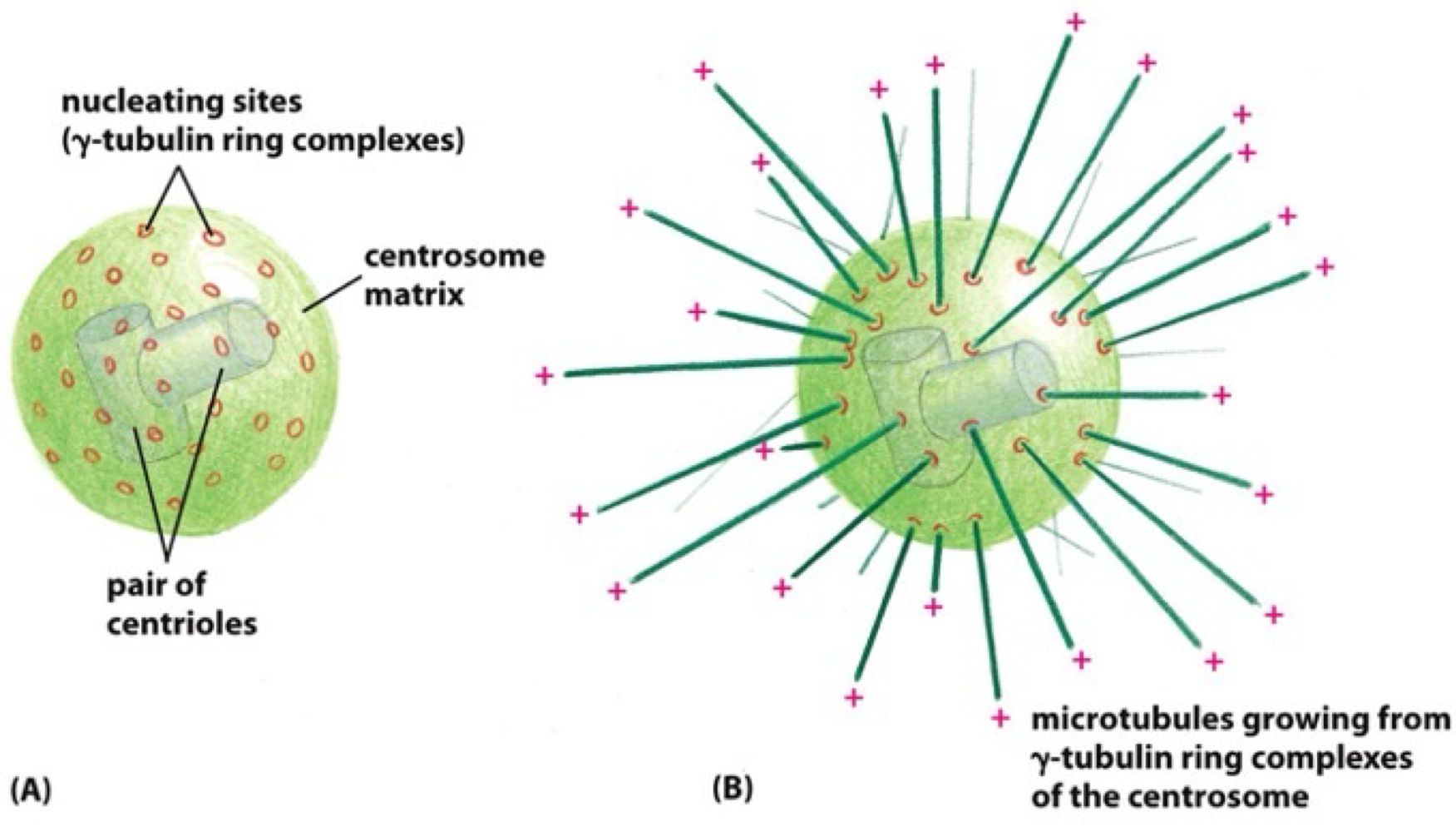
How does microtubule nucleation occur?
spontaneous MT formation requires very high concentrations of α + β tubulin heterodimers
MT nucleation is catalysed by third type of tubulin = γ tubulin
γ tubulin + accessory proteins - γ tubulin ring complex = promotes MT nucleation
γ tubulin ring complex - enriched in MT organising centre in specific intracellular location
What is the MT organising centre?
centrosome = mtoc in most cells
γ tubulin ring complex are enriched at centromere
+ end microtubule grows outwards relative to centromere
What is microtubule dynamic instability?
microtubules can both grow + shrink. requires constant energy imput - GTP
How do microtubules grow?
Addition of GTP bound tubulin dimers to growing (+) end of MT (GTP cap)
addition is faster than GTP hydrolysis to unstable GDP bound β-tubulin
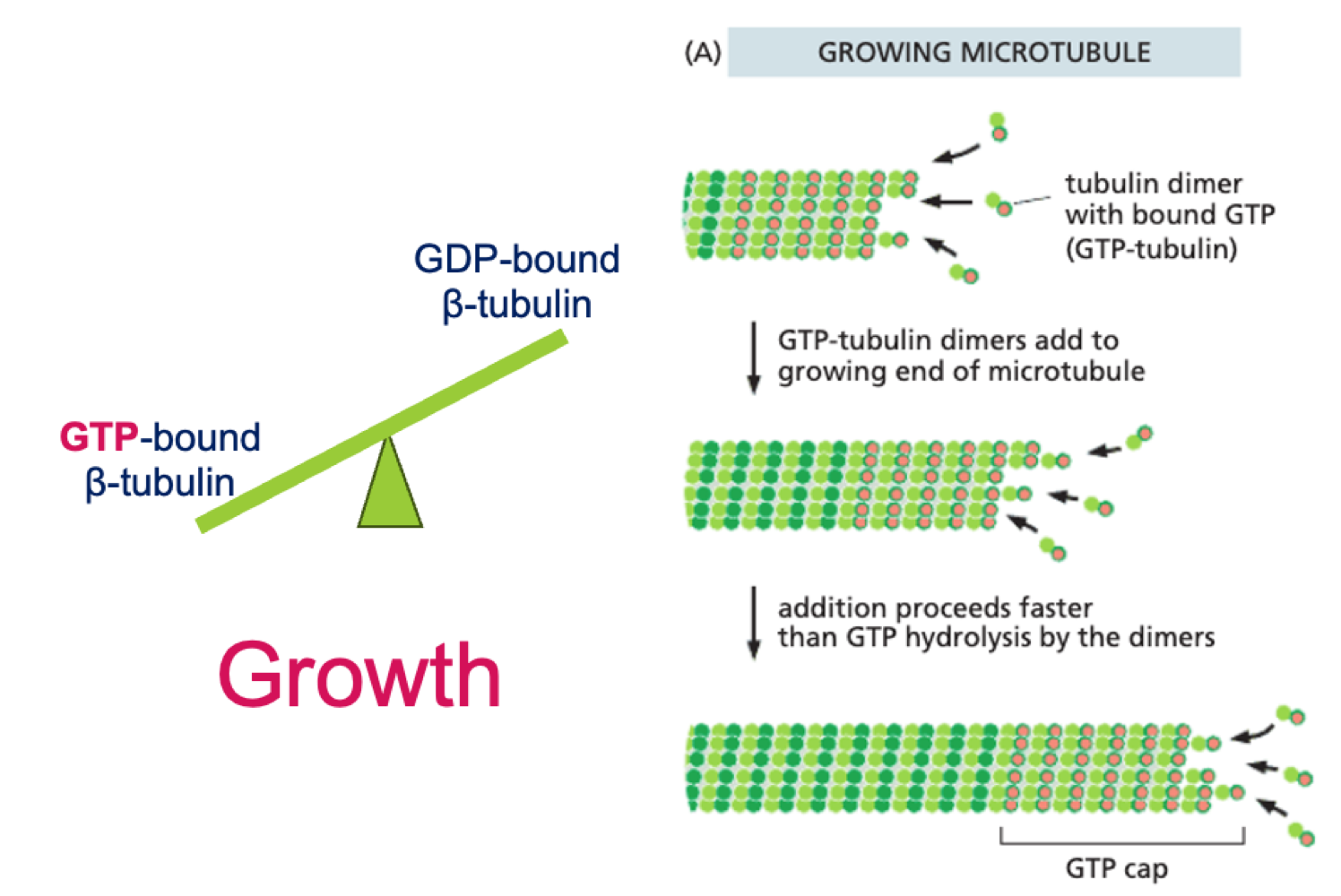
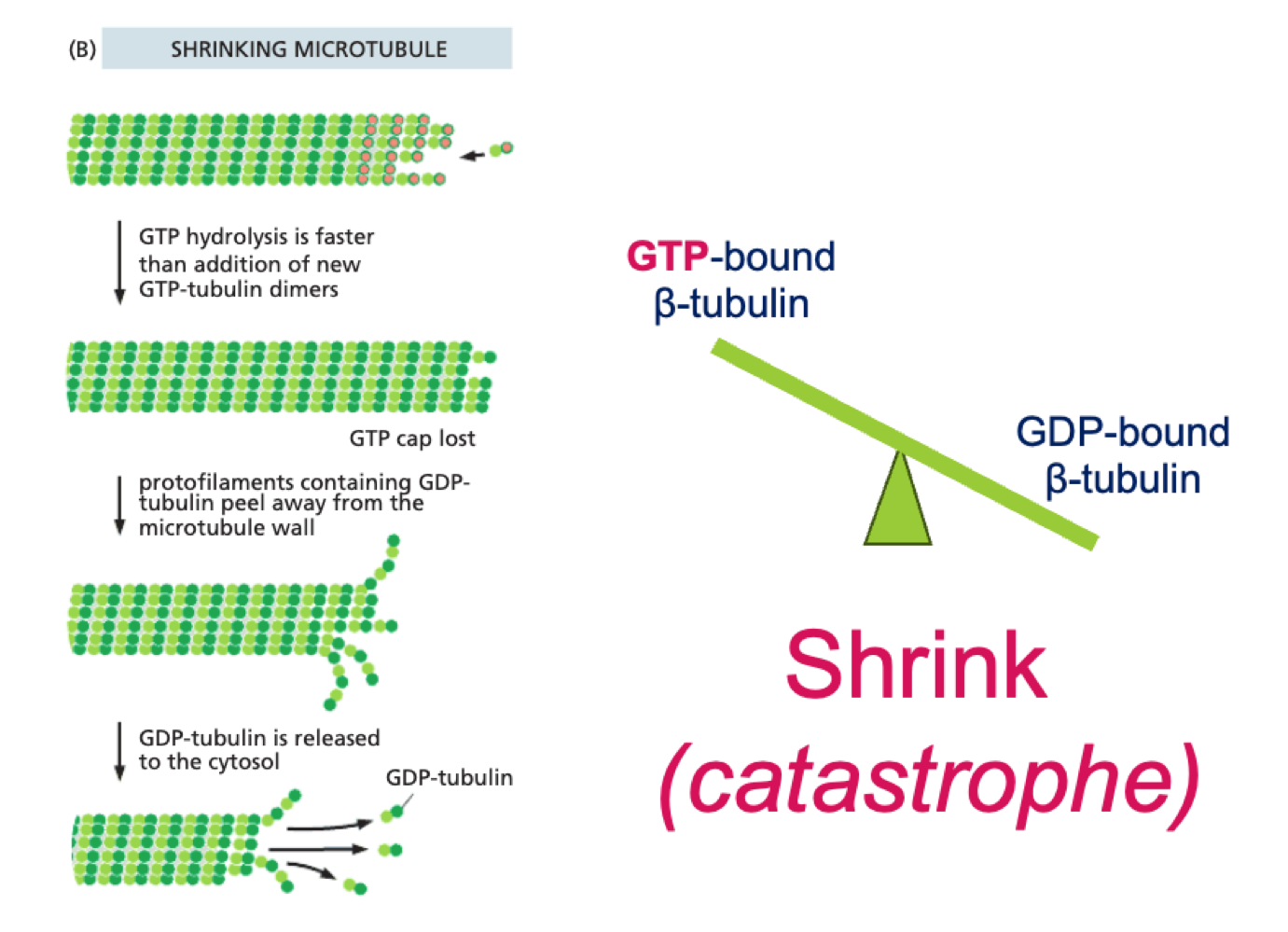
How do microtubules shrink?
GTP bound β tubulin is hydrolysed, GDP bound β tubulin makes MT unstable and leads to self-unpeeling - catastophe
What is the function of dynamic stability?
+ end of microtubule can be stabilised by attaching to another molecule or cellular structure - wont undergo dynamic instability
dynamic instability allows MT to explore cellular space
How do drugs affect dynamic instability?
Taxol (anti cancer drug) stabilises tubulin into MT lattice
prevents depolarisation
no shrinkage
results in mitotic arrest + death
Colchicine (treats gout) binds free tubulin dimers
prevents incorportation into MT - no polymerisation
results in mitotic arrest + death
What is the function of microtubules?
organisation is related to function
organelle positioning
vesicle movement
mitotic spindle
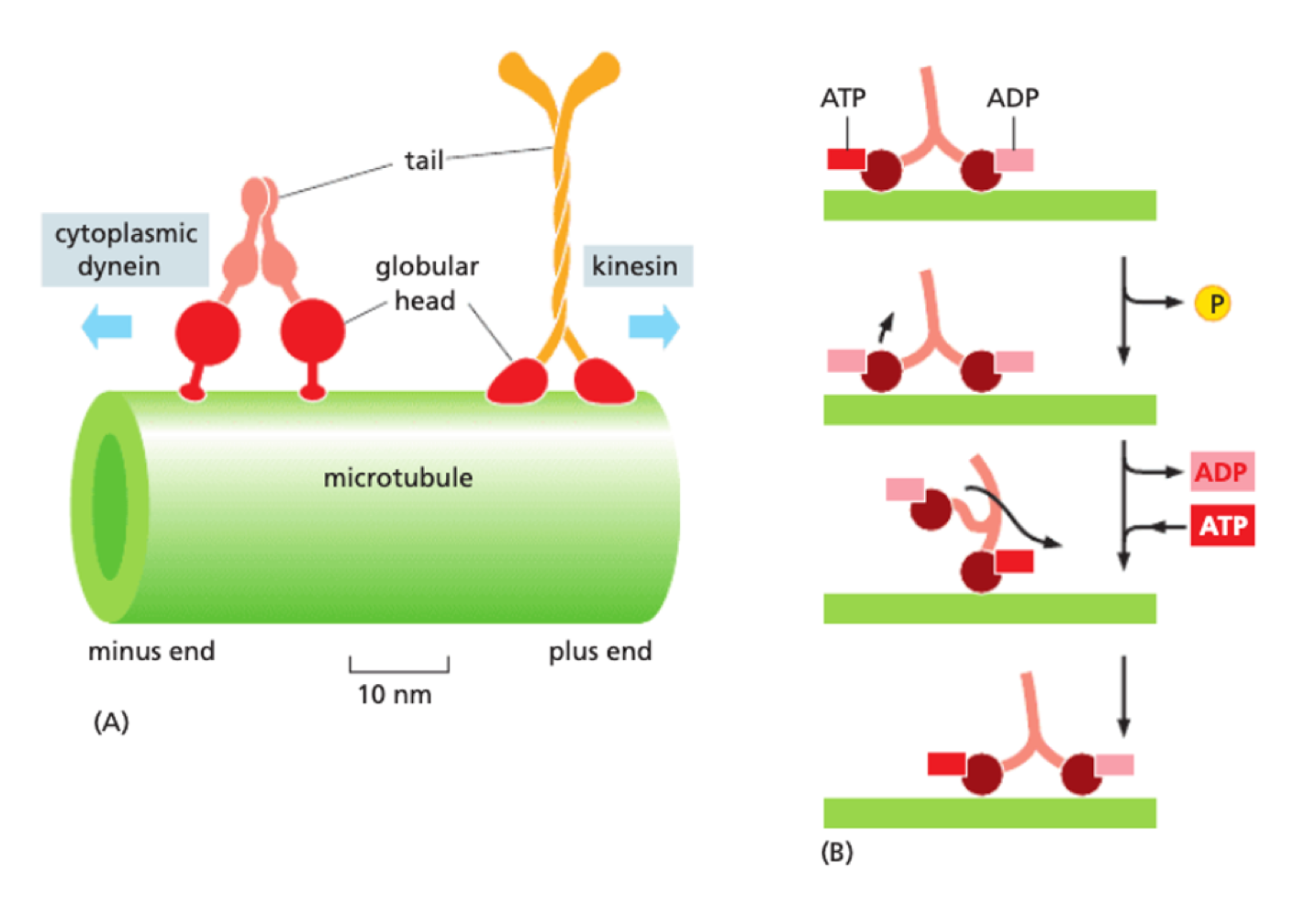
How do motor proteins aid vesicle movement?
motor proteins bind to microtubule - each type only moves in one direction + occurs in pairs
Kinesin - plus-end directed (towards + )
Dynein - minus end directed (towards - )
energy for movement from hydrolysed ATP
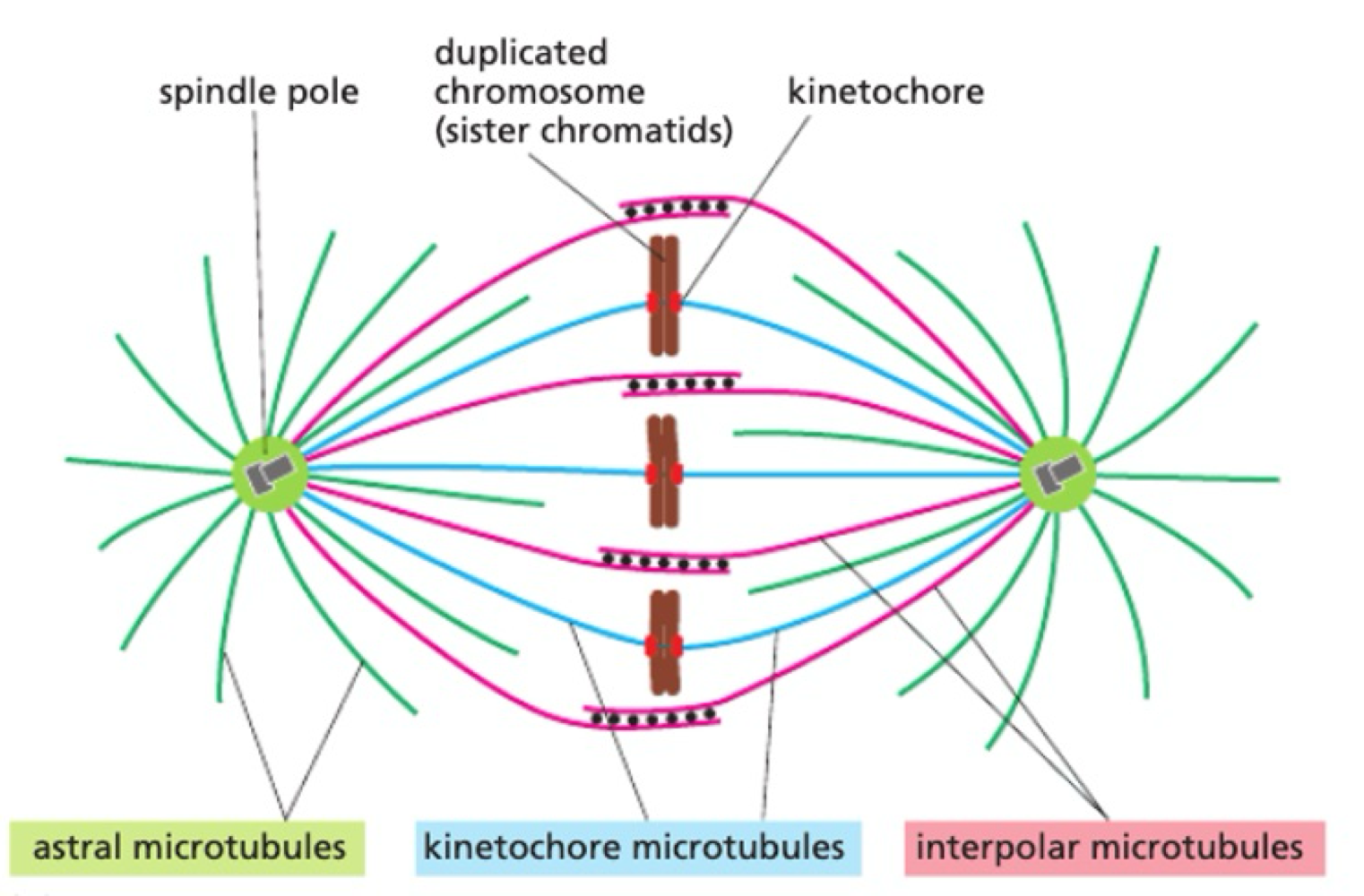
How do microtubules act as mitotic spindles?
during mitosis:
MT nucleates from centrosome
MT attached to kinetochore on chromosome = stabilised
selective stabilisation of interpolar MT through MT associated proteins
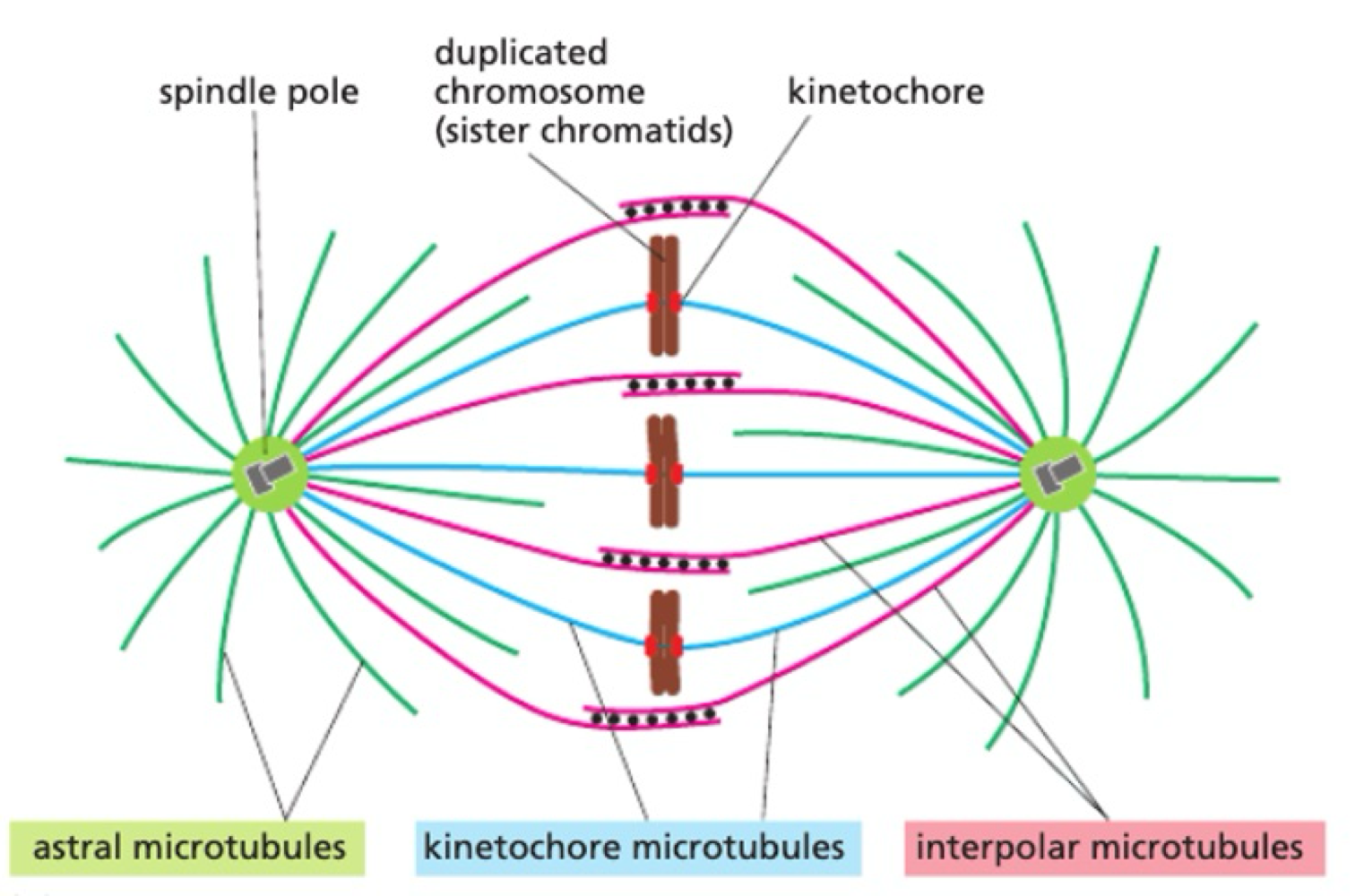
How do chromosomes move during mitosis?
pulling force = motor protein on kinetochore MT towards centrosome
pushing force = motor proteins on interpolar MT push to elongate mitotic spindle
Where do cilia + flagella originate from?
extensions from centrioles - centre of centrosome. Form base of cilia/flagella (basal bodies)
How are MT organised in cilia/flagella?
basal body = cylinder of 9 MT filaments
9” 2” arrangement - 9 outer MT doublets + 2 central MT pairs
outer MT doublets connected by nexin proteins
Dynein motor proteins bind to MT doublets - provides movement
This structure allows Active Transport System to occur = Intraflagellar Transport - for assembly + maintenance of cilia