Lecture 4: DNA repair & recombination
1/102
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
103 Terms
Proof-reading
Repair of base mismatch occurring during replication
Types of repair of neo-synthesised DNA
mismatch repair, base-excision repair, nucleotide-excision repair, direct repair
What happens when base mismatch occurs during replication?
Different geometry
Directly and immediately removed by DNA polymerase during replication
3’ → 5’ exonuclease activity of DNA pol I (or ε, δ)
How is parent strand identified?
Parental DNA is methylated in first few minutes after replication
Methylation in bacteria
Dam methylase (bacterial DNA methylase) recognises the GATC sequence
Adds the -CH3 group at the N6 positions of the Adenine (S-adenosyl methionine is the alkylating agent)
Methylation in eukaryotes
In eukaryotes, methylation occurs in the Cytosine that precedes a Guanine, in a 5’CpG
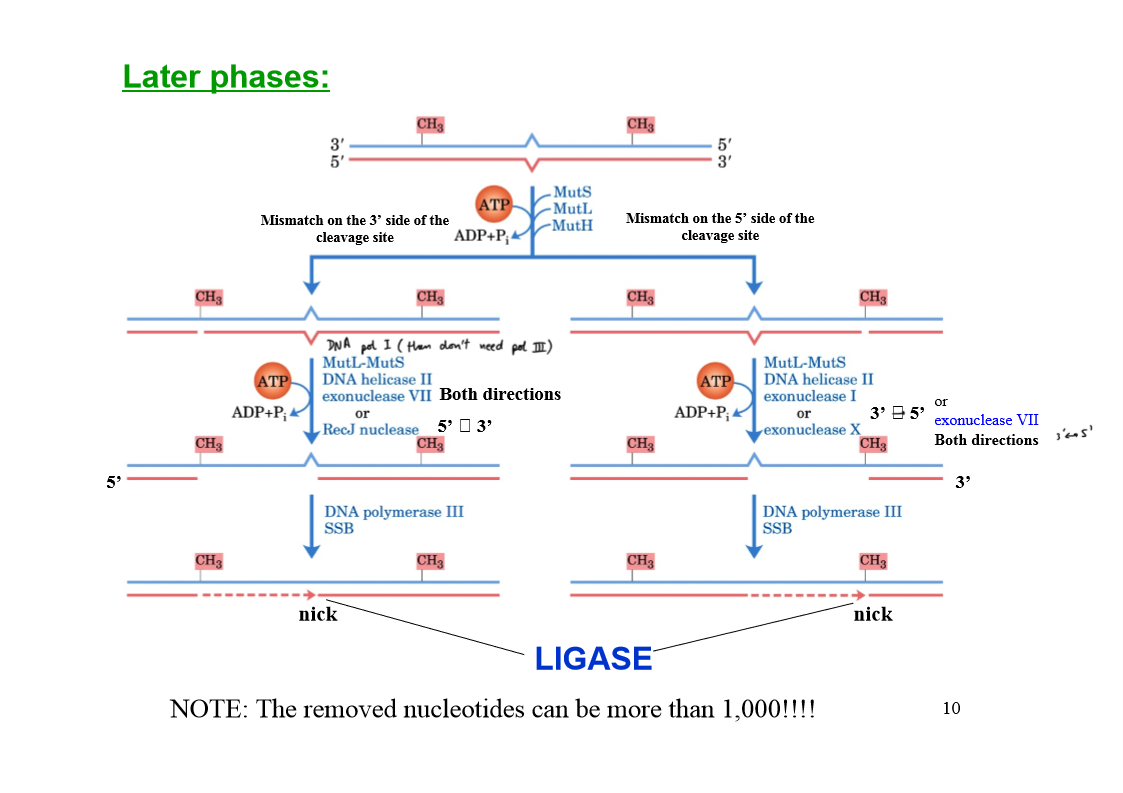
Early phases in the mismatch repair in E. coli
MutS scans DNA and forms a clamp-like complex when it encounters a lesion (complex binds to all mismatched bases, except C-C).
MutL forms a complex with MutS.
The MutSL complex slides along the DNA to find a hemimethylated GATC sequence.
MutH binds to MutL, forming MutSLH complex which moves in either direction at random.
When a hemimethylated GATC sequence is found, MutH (an endonuclease) is activated.
Activated MutH cleaves the unmethylated strand on the 5' side of the G in the GATC, marking the strand for repair.
Later phases of mismatch repair
Mismatch repair system in eukaryotes
homolog of MutS → MSH2, MSH3, MSH6
homolog of MutL → heterodimer MLH1 + PMS1
MutH → no homolog in eukaryotes
MSH2-6 heterodimers bind to….?
Single bp mismatches and less to slightly longer mismatches
MSH2-MSH3 dimer binds…?
Longer mismatches (2-6 bp) in many organisms
Base-excision DNA repair
Excision of bases recognised as DNA components
e.g. uracil which could replace cytosine by deamination and pair to adenine or hypoxanthine which could replace adenine by deamination and pair to cytosine, as a guanine
Why does DNA contain Thymine and not Uracil like RNA
Uracil spontaneously forms (slow reaction) by deamination of cytosine, therefore it would become not recognisable from other U bases, if they were “physiologically” allowed.
Thus, when U forms in the DNA it is recognised and eliminated
Enzymes involved in base-excision DNA repair of an AP site
DNA glycosylase
AP endonuclease + helicase
DNA polymerase I
DNA ligase
DNA glycosylase
Cleaves bond between base and ribose
AP endonuclease
Cuts phosphodiester bond at AP site
How many types of uracil DNA glycosylases in bacteria and humans?
Bacteria: Only 1 type
Humans: At least 4 types
UNG
Eliminates occasional U instead of T in DNA at replisome
hSMUG1
Removes any U in ssDNA during replication or transcription
TDG
Removes U paired with G by deamination by C (deaminated C is U)
MBD4
Removes T paired with G by 5-methylcytosine (deaminated 5-methyl is T)
Nucleotide-excision repair
For DNA lesions that large distortions in the helical structure of DNA
Critical pathway for the survival of all free-living organisms
Excinuclease
Multi sub-unit enzyme
Hydrolyses 2 phosphodiester bonds, one on either side of the distortion caused by the lesion
Excinuclease in E. coli features
5th bond 3’ side, 8th bond on 5’ side (+1-2 damaged nucleotides)
12-13 nucleotide fragments
Excinuclease in eukaryotes
6th on 3’ side, 22nd bond on 5’ side (+ 1-2 damaged nucleotides)
27-29 nucleotide fragments
Nucleotide-excision repair in E. coli
E. coli excinuclease → DNA helicase (13 mer) → DNA pol I → DNA ligase
Nucleotide-excision repair in humans
Human excinuclease → DNA helicase (29 mer) → DNA pol ε → DNA ligase
ABC excinuclease system subunits
UvrA, UvrB, UvrC
Mechanism of the ABC excinuclease system
UvrA dimer (ATPase) scans DNA and binds to a lesion
UvrB binds to UvrA
If a lesion is found UvrA dissociates and leaves a tight UvrB-DNA complex
UvrC binds to UvrB
UvrB makes an incision at the 5th bond on the 3’ side of the lesion
UvrC makes an incision at the 8th bond on the 5’ side of the lesion
12-13mer is removed by UvrD helicase
Gap repaired by DNA Pol I (E. coli)
Nucleotide-excision repair is the primary repair route for…?
cyclobutane pyrimidine dimers, 6-4 photoproducts, benzopyrene-guanine
How is nucleotide-excision repair different in eukaryotes than bacteria?
Similar mechanism but at least 16 proteins with no similarity to ABC excinuclease system are involved
Direct repair
Lesion is corrected in place
How is O6-methylguanine repaired? What type of reaction is it?
Direct repair by O6-methylguanine methyltransferase
-CH3 removed → Guanine formed
Non-enzymatic reaction (protein methylation permanently inactivates the methyltransferase)
In which situations is DNA repair not possible by proofreading, mismatch repair, base-excision repair, nucleotide-excision repair or direct repair?
Unrepaired lesion halts replication fork and generates a ssDNA
Strand break halts the replication fork (one of the arms is lost) and the fork collapses
What can be used to correct:
Unrepaired lesion halts replication fork and generates a ssDNA
Strand break halts the replication fork and the fork collapses
DNA recombination
Error-prone TLS (trans-lesion DNA synthesis)
TLS is part of _________ in bacteria
SOS response
What proteins are involved in TLS?
UvrA, UvrB, UmuC, UmuD
TLS in bacteria
UmuD cleaved in SOS-regulated process to UmuD’
UmuD’ binds to UmuC and RecA to form DNA pol V
DNA pol V is made of…?
UmuD’2-UmuC-RecA
DNA pol V function
Can replicate past many of the DNA lesions that would normally block replication
Proper base pairing is impossible → error-prone system (desperate strategy)
Like DNA pol V, what other enzyme is also very error-prone?
DNA pol IV
What is the error rate of DNA pol IV and V
Reduces fidelity to 1 error in 1000 nt
Which enzymes involved in TLS in mammals?
Low-fidelity polymerases:
DNA polymerase η (all eukaryotes)
DNA polymerases β, ι, λ
DNA polymerase η
Promotes TLS across cyclobutane T-T dimers
Few mutations occur since the enzyme preferentially inserts A-A
DNA polymerases β, ι, λ
After the action of glycosylase and AP endonuclease, they remove the abasic site and randomly fill the very short gap.
The very short length of DNA synthesised minimises the mutation rate
Define DNA recombination
The rearrangement of genetic information within and among DNA molecules
Types of DNA recombination
Homologous genetic recombination
Site-specific recombination
DNA transposition
Homologous genetic recombination
Involves genetic exchanges between any 2 DNA molecules or segments that share an extended region of nearly identical sequence
Includes a process to repair ds breaks in DNA (NHEJ)
Site-specific recombination
Exchanges occur only at specific DNA sequences
DNA transposition
Involves a short segment of DNA capable of moving from one location to another (jumping genes)
Homologous recombination is primarily a DNA repair process in …?
bacteria (recombination DNA repair)
Homologous recombination purposes
Reconstruction of replication forks (stalled or collapsed)
Increasing genetic diversity during conjugation
Recombinational DNA repair at a collapsed replication fork
The 5’ ending strand at the break is degraded
ss 3’-extension is created
3’-extension invades and pairs with its complementary strand in the adjacent duplex
Migration of the branch creates a Holliday intermediate
Specialised nucleases resolve the Holliday intermediate
Ligation restores a viable replication fork and replication resumes
Holliday intermediate
A branched nucleic acid structure that contains 4 double-stranded arms joined together in several possible conformations
What is responsible for homologous recombination in E. coli?
RecBCD nuclease/helicase system
RecD
5’ → 3’ helicase
RecB
3’ → 5’ helicase, nuclease (degrades both DNA strands as they unwind)
Describe homologous recombination in E. coli
Helicases (RecB and RecD) halt at CHI sequences
CHI sequence binds tightly to RecC
Creation of ss 3’-extension
Degradation of 3’ ending strand is greatly reduced
Unwinding and degradation of 5’ ending strand is increased
RecA recombinase is loaded
Promotes strand invasion + strand exchange reactions
RuvAB complex promotes branch migration
Formation of Holliday intermediate
RuvC specialised nucleases cleave and resolve the Holliday intermediate
Nicks are sealed by DNA ligase
CHI sequence
5’-GCTGGTGG-3’
RuvA
Binds to the Holliday intermediate
RuvB
Hexameric, translocase
2 RuvB hexamers bind to opposite arms of the Holliday intermediate
Propel DNA outward in a reaction coupled to ATP hydrolysis
The branch thus moves
RuvC
Binds to RuvAB and cleaves the Holliday intermediate on opposite side of the junction
The 2 contiguous DNA arms remain in each product
What happens after the replication fork reassembles?
Origin-independent restart of replication
Replication restart primosome
PriA, PriB, PriC, DnaT, DnaC, DnaB, DnaG
Describe origin-independent restart of replication
4 proteins (PriA, PriB, PriC, DnaT) act with DnaC to load DnaB helicase onto the reconstructed replication fork
DnaG primase synthesises an RNA primer
DNA pol III reassembles on DnaB to restart DNA synthesis
Functions of homologous recombination in eukaryotes
Repair of several types of DNA damage
Transient physical link between chromatids promoting orderly segregation of chromosomes at the first meiotic division
Enhancement of genetic diversity in a population
Where does homologous recombination take place in eukaryotes?
Chiasmata
How are chiasmata formed?
In prophase I, just before the first meiotic division, the 2 sets of chromatids (each have replicated dsDNA molecule) align to form tetrads, held together by covalent links at homologous junctions (chiasmata).
How is proper segregation ensured in dividing cells?
Transient associations between homologs creates a tension that allows proper segregation when spindle fibers pull them towards the poles of dividing cells in the first meiotic division
What is NHEJ and when is it needed?
Non-homologous end joining
Ds breaks occurring when recombinational DNA repair is not feasible
e.g. when the break occurs in phases in which DNA is not replicating + no sister chromatids
Disadvantages of NHEJ
Highly mutagenic; smaller genomes cannot afford it → they privilege homologous recombination
Does not preserve the original DNA sequence
Ku70-Ku80
Heterodimer binding the DNA ends (scaffold to assemble other molecules)
DNA-PKcs
Protein kinase
Artemis
Nuclease
P-Artemis
Endonuclease
Describe NHEJ
DNA ends bound by Ku70-Ku80
Ku70-Ku80 recruits DNA-PKcs and Artemis
The 2 DNA ends are synapsed (held together)
DNA-PKcs autophosphorylates and phosphorylates Artemis
P-Artemis is activated, and removes 5’- or 3’-ss extensions or hairpins at the ends
A helicase separates the 2 ends and strands from different ends are annealed at regions of short complementarity.
Artemis removes any unpaired DNA.
Small gaps are filled by either DNA Polymerase μ or λ
The nicks are sealed by a protein complex composed of XRCC4, XLF and DNA ligase IV
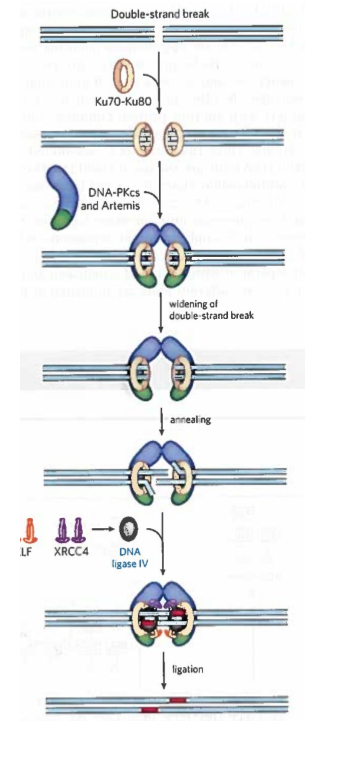
NHEJ consequence
Broken ends are kept close together by chromatin structures to avoid joining far apart areas; could lead to deleterious rearrangements
Where does site-specific recombination occur?
In every cell
Role of site-specific recombination
Gene expression regulation
Programmed DNA rearrangements in embryo development
Replication cycle of viral, plasmidic DNA
Site-specific recombination requires…?
Recombinase (2 classes: Tyr or Ser in active site)
Short (20-200 bp), unique DNA sequences (recombination site)
One or more auxiliary proteins (timing of reaction)
Tyr-class recombinases
2 separate recombinases on recombination site within same or on different DNA molecules
One strand in each site is cleaved at a specific point within the site.
Recombinase covalently links to the DNA (phospho-tyrosine bond).
Transient protein-DNA linkage preserves the phosphodiester bond lost in DNA cleavage, so ATP not necessary in following steps.
Cleaved DNA is re-joined to new partners at the expense of P-Tyr bond (formation of Holliday intermediate)
Isomerization: Process repeated at a second point within each of the 2 rec sites
Recombinases act as both ______ and ______
site-specific endonuclease and ligase
Ser-class recombinases
Both strands of each recombination site are cut simultaneously and rejoined to new partners without Holliday intermediates
In both Tyr- and Ser- classes…?
The exchange is always reciprocal, precise, and regenerates the recombination sites when completed
What happens if 2 recombing sites align in the opposite orientation during the recombinase reaction?
Same DNA molecule: inversion
What happens if 2 recombing sites align in the same orientation during the recombinase reaction?
1 DNA molecule: deletion
2 DNA molecules: insertion
Transposons
Segments of DNA that can move (“jump”) from one place (donor site) to another in the same or a different (target site)
Where are transposons found?
In all cells
Is transposition a random process?
More or less, but it is tightly regulated (it could kill the cell)
Is DNA sequence homology required for transposons?
No
What are the classes of transposons in bacteria?
Insertional sequences (simple transposons)
Complex transposons
Insertional sequences
Sequence required for transposition + genes for transposases
Complex transposons
Also contain one or more genes (relevance for the transmission of antibiotic resistance among bacteria)
Transposase definition + mechanism
Enzyme promoting transposition
Makes staggered cuts in the target site
Transposase binding sites
Short repeated sequences (short terminal repeats, 5-10 bp)
What happens when a transposon is inserted into target site?
Transposition results in the duplication of flanking DNA sequence at target site
Direct transposition
Cut on each side of transposon and excision (leaves ds break that needs repair at the donor site)
Staggered cut at the target site → transposon is inserted and DNA replication fills the gaps to duplicate the target-site sequence
Replicative transposition
Transposon is replicated (a copy is left at the donor site)
An intermediate cointegrate forms
Cointegrate consists of the donor region covalently linked to DNA at the target site and contains 2 copies of the transposon, with the same relative orientation
Possible site-specific recombination in cointegrate intermediate
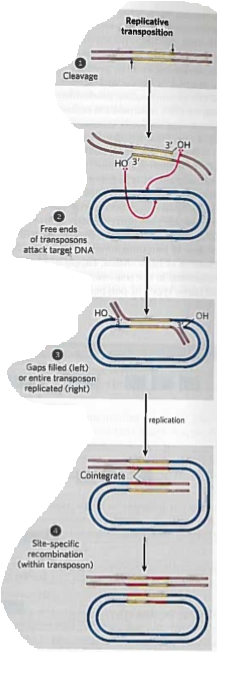
Eukaryotic transposons
Structurally similar to bacterial transposons
Similar transposition mechanisms
In some cases transposition involves an RNA intermediate
Immunoglobin genes assemble by…?
Recombination