Principles of Cell and Systemic Physiology
1/41
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
42 Terms
The Plasma Membrane
A thin phospholipid bilayer studded with membrane proteins enclosing each cell
Confer selective permeability to ions, glucose, and other molecules
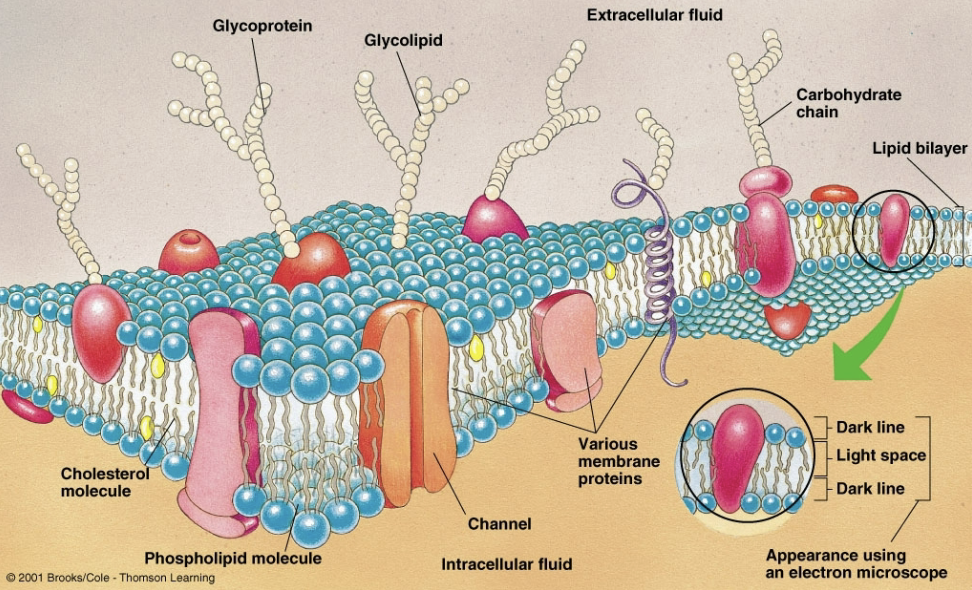
Membrane Proteins
Imbedded into the plasma membrane
Channels and carries to transport molecules and ions into and out of the cell
Receptors to signal responses
Form adhesions and junctions
The Nucleus
Membrane bound organelle containing the genetic material
Is the sight of replication and transcription (produces mRNAs that are exported to the cytoplasm)
The Cytoplasm
Portion of the cells interior not occupied by the nucleus
Contains cytosol, organelles, and the cytoskeleton
The Cytosol
Semiliquid portion of the cytoplasm
Enzymatic regulation of intermediate metabolism
Ribosomal protein synthesis
Storage of fat and glycogen
Ribosomes
Site of protein synthesis (translation)
Found studded on the endoplasmic reticulum (ER) or free in the cytosol
Organelles
Membrane-enclosed structures that carry out specific functions
Five main types similar in all cells
Endoplasmic Reticulum
Golgi Complex
Lysosomes
Peroxisomes
Mitochondria
Endoplasmic Reticulum (organelle)
Continuous fluid filled network of membranous tubules
Rough ER: membrane covered with ribosomes
Smooth ER: membrane lacking ribosomes
Golgi Complex (organelle)
Processes raw material into finished products
Directs products to their destination
Lysosomes (organelle)
Membrane-enclosed sacs containing hydrolytic enzymes
Digest debris by fusing with intracellular vesicles often derived from endocytosis
Peroxisomes (organelle)
Membrane-enclosed sacs containing oxidative enzymes which act to remove hydrogen from toxic molecules
Detoxify free radicals
Mitochondria (organelle)
Responsible for aerobic metabolism and the production of cellular energy (ATP)
Produces ATP from glucose or fatty acids (amino acids in extreme cases)
Aerobic Metabolism (mitochondria)
Glycolysis: occurs in cytosol, no oxygen required, yields 2 ATP
TCA Cycle: occurs in mitochondria, no oxygen required, yields 2 ATP
Electron Transport: occurs in mitochondria, oxygen required, yields 28-32 ATP
How do the endoplasmic reticulum and Golgi complex work together?
The ER and Golgi complex use vesicle-based systems, budding and fusion, to sort new proteins to either the plasma membrane, outside the cell (soluble proteins released by exocytosis), or lysosomes
Proteins made in the ER are never part of the cytoplasm, they are contained in the lumen
Once proteins are synthesized on ribosomes, they stay inside the endomembrane system
Where do organelles (besides lysosomes) and the cytoplasm get their proteins?
The mitochondria makes a few proteins from their own mini genome and transcription/ translation apparatus
Other organelles and the cytoplasm get their proteins from free ribosomes
The Cytoskeleton
Protein network for structural support, transport, and cellular movement
The major components
Microtubules
Microfilaments
Intermediate Filaments
Microtubules (cytoskeleton)
Dynamic polymers of tubulin
Form highways for movement of transport vesicles via kinesin and dynein motor proteins, and cilia and flagella for generating movements
Microfilaments (cytoskeleton)
Dynamic polymers of actin
In association with myosin (motor protein), they produce cellular contraction
Intermediate Filaments (cytoskeleton)
Longer proteins produced by an array of different genes
Provide support for components subject to mechanical stress
What does complex multicellular life require?
Many different types of cells specialized for different tasks
Differential gene expression is the proximate cause
All cell types contain the same DNA, but express unique subsets for any given cell type
Levels of Organization
Cell
Tissue
Organ
Organ System
Organism
Tissue (levels of organization)
Aggregate of cells and extracellular material
Four types
Muscle: contraction
Nervous: electrochemical signals
Connective: structural support
Epithelial: exchange
Exocrine: external secretion
Endocrine: internal secretion
Organ (levels of organization)
Two or more primary tissues organized to perform a function
Organ System (levels of organization)
Organs working together to perform a function to maintain homeostasis
Homeostasis
Dynamic maintenance of a stable internal (extracellular) environment within the organism
Essential to the survival of each cell
Requires continual exchange of material between the inter and extracellular spaces
Each organ system contributes by counteracting changes of internal environments
Intrinsic (homeostasis)
Local control system built into an organ
Extrinsic (homeostasis)
External control system outside of an organ permitting coordinated regulation of several organs
Negative Feedback (homeostasis)
Change in a controlled variable triggers a response that opposes the change- return to the ‘normal’ state, maintain homeostasis
Sensor, set point, integrator, effector
Sensor (negative feedback)
Mechanism to detect the controlled variable
Constant
Set Point (negative feedback)
The desired value of the variable
Integrator (negative feedback)
Compares the sensor’s input with the set point
Notices the direction and magnitude of change
Activates or inhibits the effector
Effector (negative feedback)
Adjusts the value of the controlled variable
Activated or inhibited by the integrator
Positive Feedback (homeostasis)
Reinforces the change in a controlled variable
Occurs very rarely
Extracellular Chemical Messengers
Specific ligand-receptor interactions
Ligand typically activates the receptor
Cellular response depends on the molecular identity of the receptor
Three types: hormonal, paracrine, synpatic
Hormonal (extracellular chemical messengers)
Releases signal into blood stream, which goes body wide
Exposed to all cells but only activates those with the receptor
Paracrine (extracellular chemical messengers)
Cell releases signaling molecules which are detected by cells around it which have the receptor
Cells in the same tissue
About 10-100 microns
Synaptic (extracellular chemical messengers)
Signal released into the extracellular space and is detected by the postsynaptic cell
About 1 micron
Detected at one part of one cell
Extracellular Chemical Messenger Receptors
Specific ligand-receptor interactions, typically the ligand activates the receptors
Cellular response depends on the molecular identity of the receptor
Four types: nuclear, GPCRs, enzyme-linked, ionotropic
Nuclear Receptor (extracellular chemical messenger receptor)
Intracellular
Chemical messenger/ molecule diffuses through the plasma membrane and binds to a nuclear receptor protein
The activated complex moves into the nucleus and binds to the regulatory region of the target gene and activates transcription
GPCR (extracellular chemical messenger receptor)
Chemical messenger binds to the cell surface receptor, which activates a G-protein cascade, leading to a sequence of phosphorylation events that alter the shape and function of preexisting proteins and bring about a cellular response
Enzyme-Linked Receptor (extracellular chemical messenger receptor)
Cell surface receptors that bind their ligand and initiate an enzymatic cascade
Inotropic Receptor (extracellular chemical messenger receptor)
Chemical messenger, a neurotransmitter, binds the receptor and opens the ion channel, allowing ions to flow down their electrochemical gradient