Biochemistry
1/400
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
401 Terms
Oxidoreductases
Catalyze oxidation-reduction reactions (transfer of electrons); Often have electron-carrying cofactor like NAD+ or NADP+; Enzymes with dehydrogenase or reductase in their names
Transferases
Catalyze the movement of a functional group from one molecule to another; I.e. Kinases (transfer of a phosphate group)
Hydrolases
Catalyze the breaking of a compound into two molecules using the addition of water; Many named for their substrate; I.e. Phosphatase cleaves a phosphate group from another molecule
Lyases
Catalyze the cleavage of a single molecule into two products; AKA synthases when catalyze reverse reaction
Isomerases
Catalyze the rearrangement of bonds within a molecule; Catalyze reactions between stereoisomers and constitutional isomers
Ligases
Catalyze addition or synthesis reactions, generally between large similar molecules, and often require ATP; Nucleic acid synthesis and repair
Endergonic Reaction
Requires energy input
Exergonic Reaction
Energy is given off
Cofactors (Coenzymes)
Nonprotein molecules that tend to be small in size so they can bind to the active site of the enzyme and participate in the catalysis of the reaction, usually by carrying charge through ionization, protonation, or deprotonation
Apoenzymes
Enzymes without their cofactors
Holoenzymes
Enzymes containing their cofactors
Prosthetic Groups
Tightly bound cofactors or coenzymes that are necessary for enzyme function
Michaelis-Menten Plot
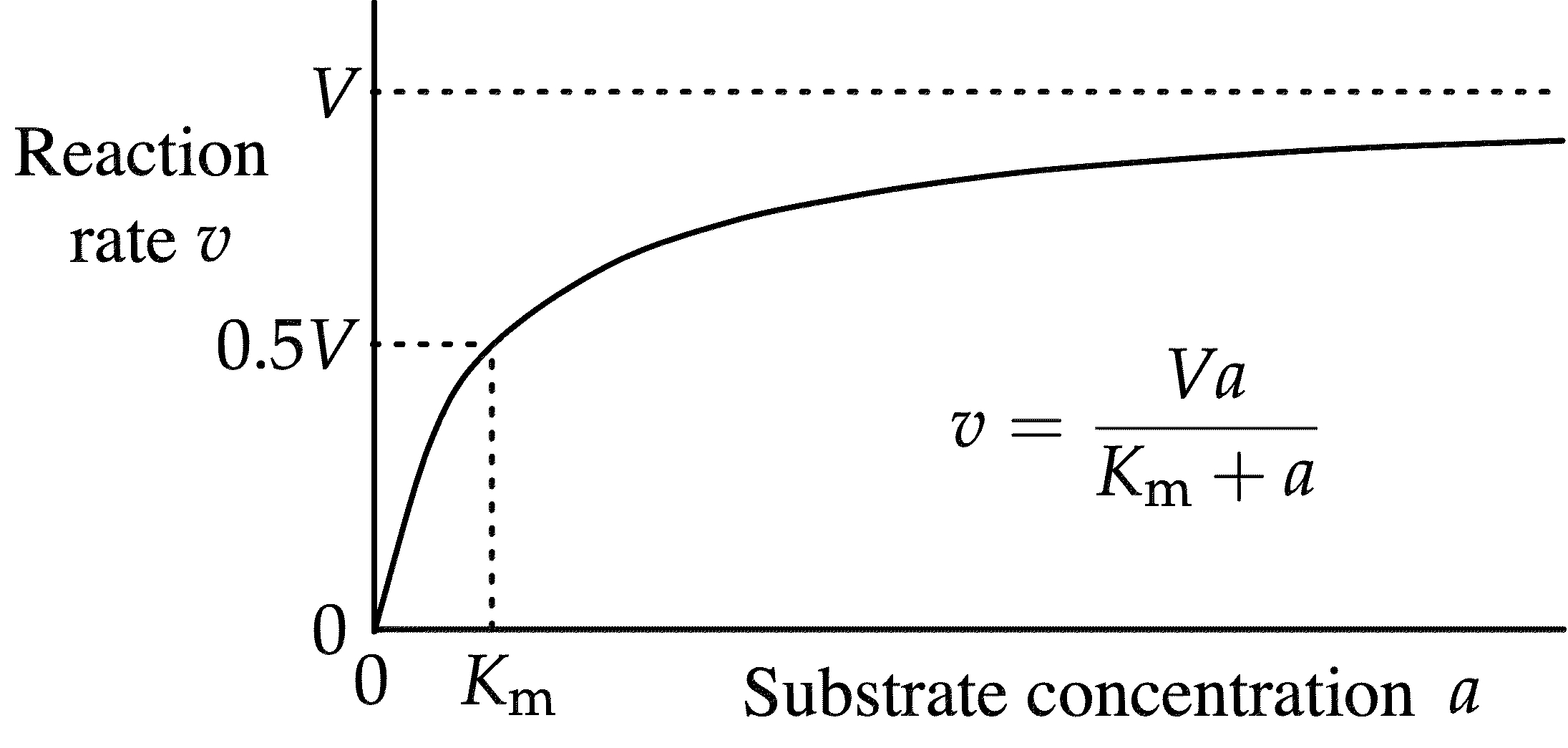
Michaelis-Menten Equation (and Vmax)
Vmax = [E]kcat
![<p>V<sub>max</sub> = [E]k<sub>cat</sub></p>](https://knowt-user-attachments.s3.amazonaws.com/06508ba6-b50f-4d7e-aea0-2098d591042f.webp)
Catalytic Efficiency
kcat/Km
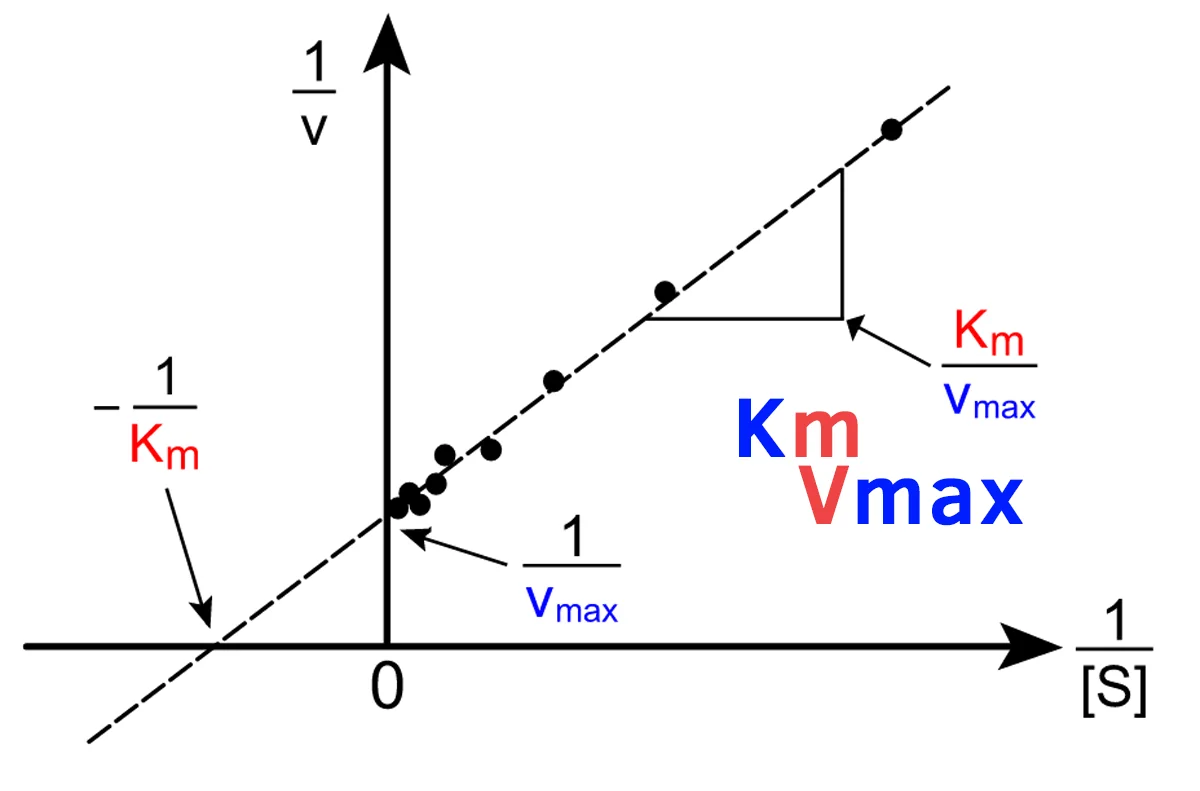
Lineweaver-Burk Plot
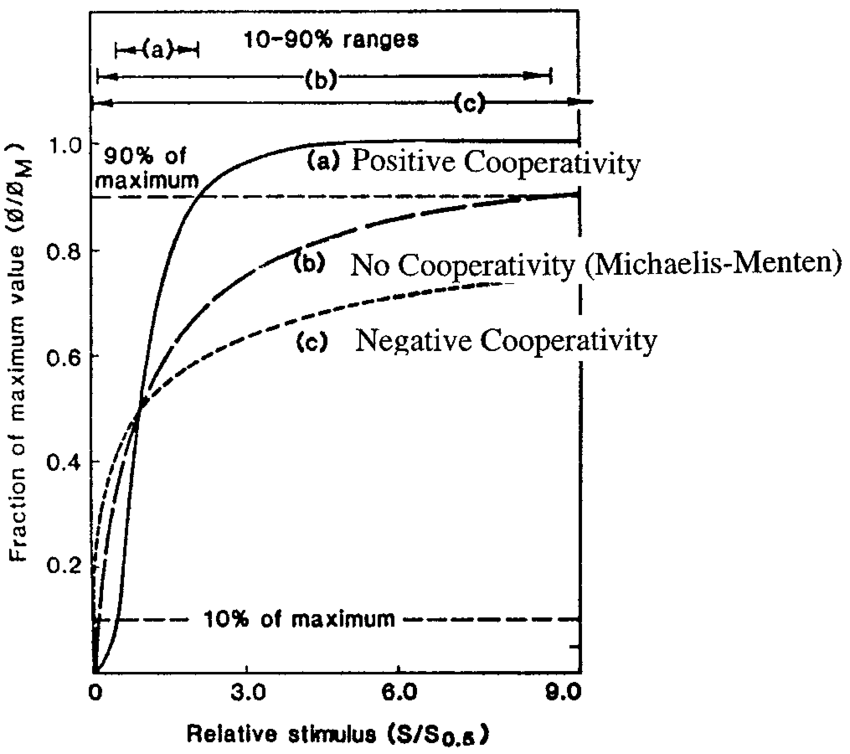
Positive Cooperativity
Hill’s coefficient > 1; I.e. After one ligand is bound the affinity of the enzyme for further ligands increases
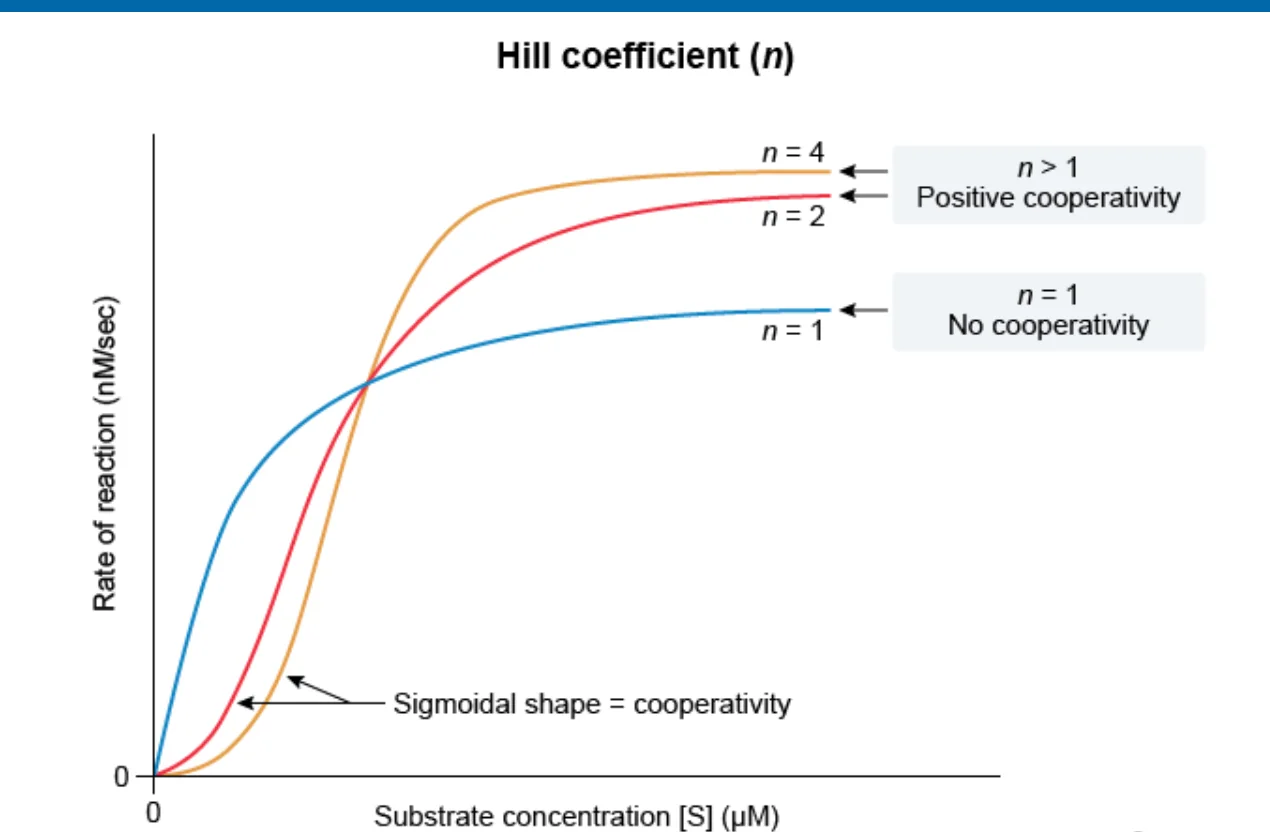
Negative Cooperativity
Hill’s coefficient < 1; I.e. After one ligand is bound the affinity of the enzyme for further ligands decreases
Competitive Inhibition
Inhibitor binds to enzyme active site; Overcome by adding more substrate so that the substrate-to-inhibitor ratio is higher; No effect on Vmax, increases Km
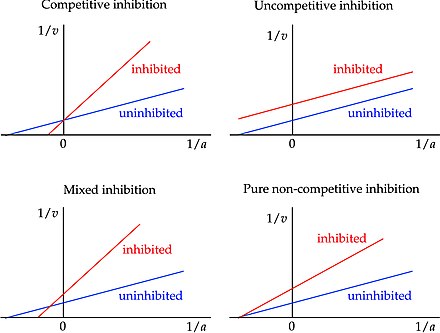
Noncompetitive Inhibition
Binds to an allosteric site on the enzyme to induce a change in enzyme conformation; Binds equally well to the enzyme and ES complex; Decreases Vmax, no effect on Km
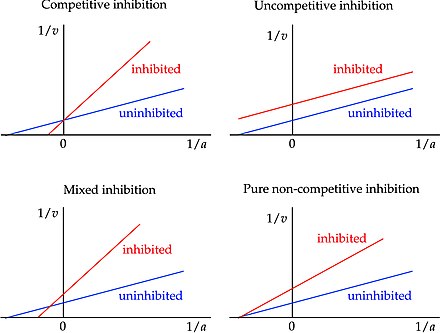
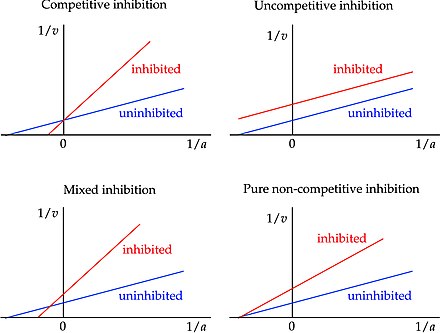
Mixed Inhibition
When an inhibitor can bind to either the enzyme or the ES complex, but has a different affinity for each; Bind at an allosteric site; Alters Km, decreases Vmax
Uncompetitive Inhibition
Bind only to the ES complex and essentially lock the substrate in the enzyme, preventing its release; Bind at an allosteric site; Lowers Km and Vmax
Allosteric Enzymes
Alternate between an active and an inactive form that cannot carry out the enzymatic reaction
Collagen
Structural protein that has characteristic trihelical fiber and makes up most of the extracellular matrix of connective tissue; Found throughout the body and is important in providing strength and flexibility
Elastin
Structural protein that is an important component of the extracellular matrix of connective tissue; Primary role is to stretch and then recoil like a spring, which restores the original shape of the tissue
Keratins
Structural intermediate filament proteins found in epithelial cells; Contribute to the mechanical integrity of the cell and also function as regulatory proteins; Primary protein that makes up hair and nails
Actin
Structural protein that makes up microfilaments and the thin filaments in myofibrils; The most abundant protein in eukaryotic cells; Have a positive and a negative side, which allow motor proteins to travel unidirectionally along this protein, like a one-way street
Tubulin
The structural protein that makes up microtubules, which are important for providing structure, chromosome separation in mitosis and meiosis, and intracellular transport with kinesin and dynein; Has a negative end usually located adjacent to the nucleus, whereas the positive end is usually in the periphery of a cell
Myosin
Primary motor protein that interacts with actin; The thick filament in a myofibril; Can be involved in cellular transport; Each subunit has a single head and neck; Movement at the neck is responsible for the power stroke of sarcomere contraction
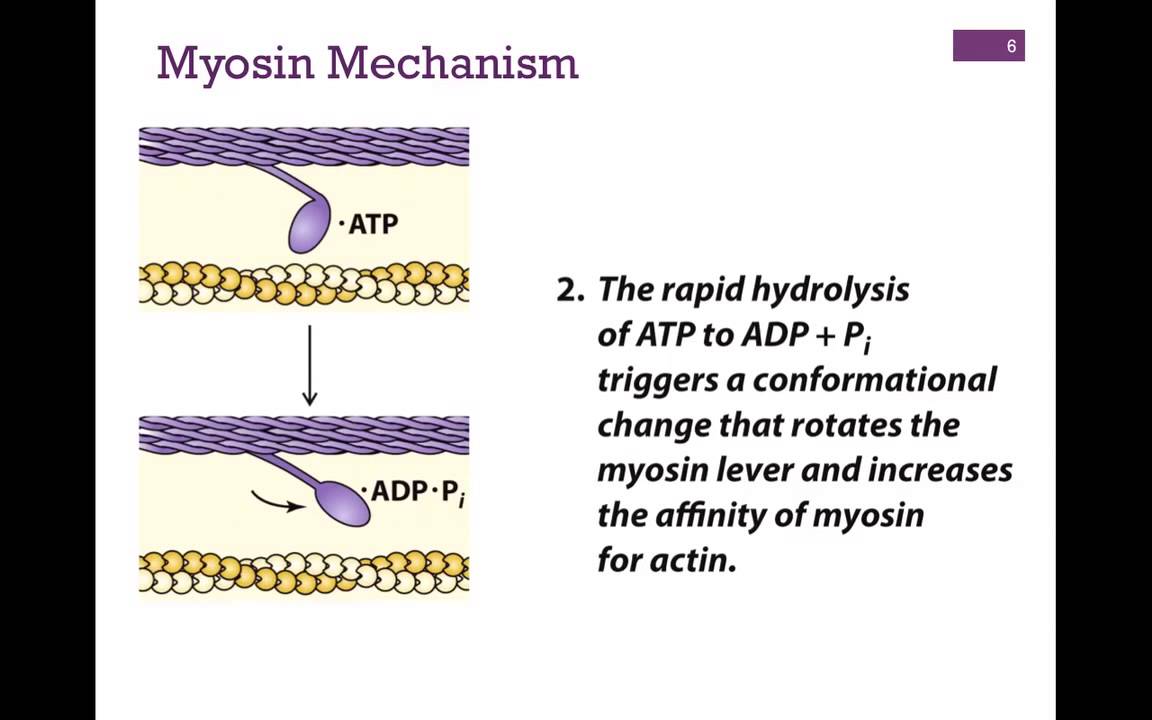
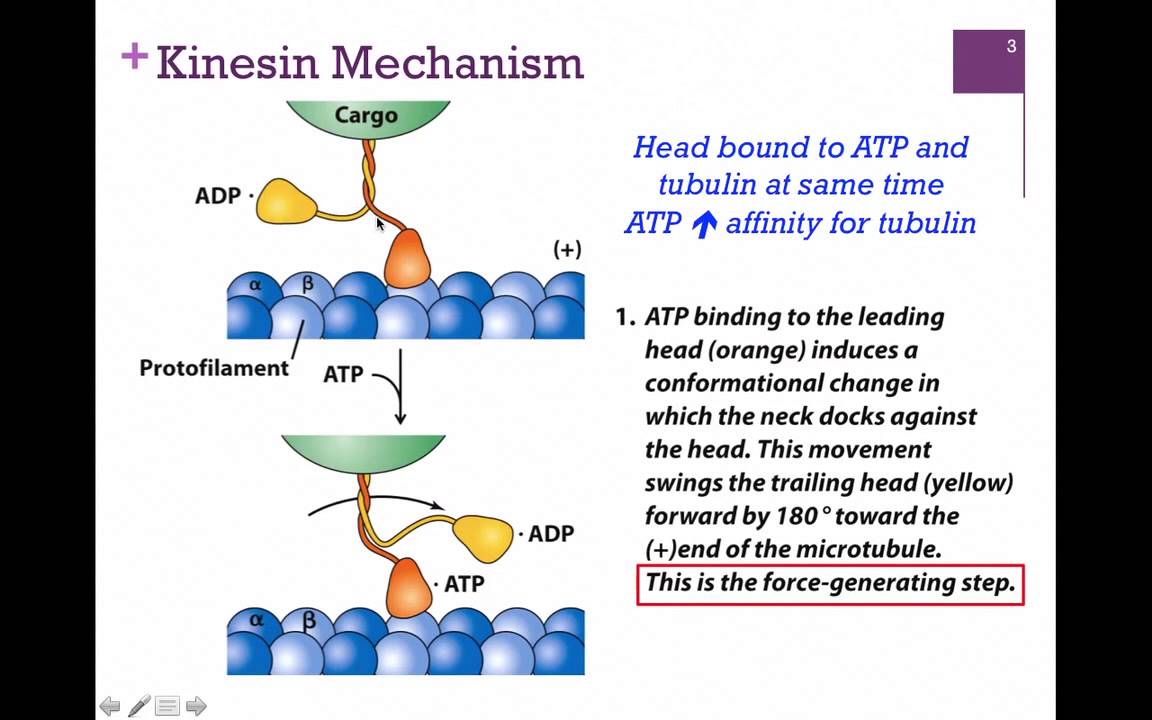
Kinesin
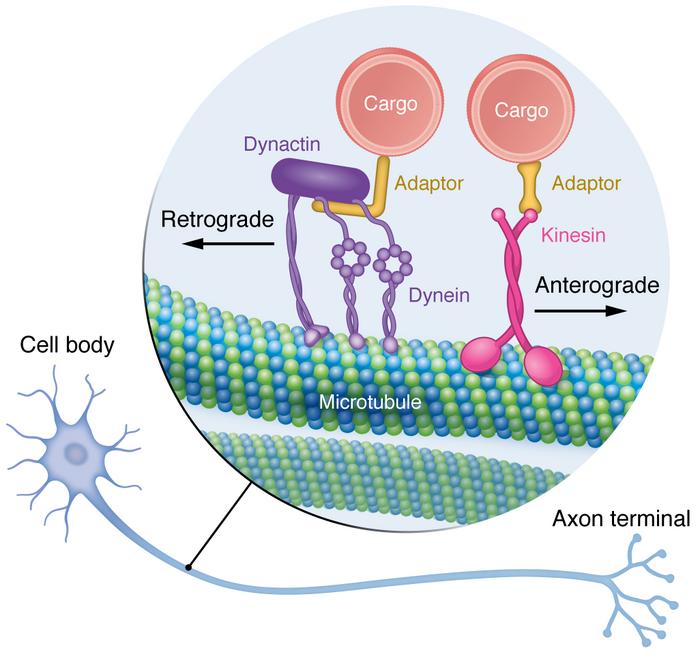
One of two motor proteins associated with microtubules; Has two heads, one head attached to tubulin at all times; Plays a key role in aligning chromosomes during metaphase and depolymerizing microtubules during anaphase of mitosis; Brings vesicles toward the positive end of the microtubule
Dynein
One of two motor proteins associated with microtubules; Has two heads, one head attached to tubulin at all times; Involved in the sliding movement of cilia and flagella; Brings vesicles toward the negative end of the microtubule
Binding Proteins
Proteins that transport or sequester molecules by binding to them; Hemoglobin, calcium-binding proteins, DNA-binding proteins, etc.; Has an affinity curve for its molecule of interest
Cell Adhesion Molecules (CAMs)
Proteins found on the surface of most cells and aid in binding the cell to the extracellular matrix or other cells; All are integral membrane proteins; Three types: Cadherins, integrins, and selectins
Cadherins
Group of glycoproteins that mediate calcium-dependent cell adhesion; Often hold similar cell types together; Type of CAM
Integrins
Group of proteins that all have two membrane-spanning chains called α and β which are important in binding to and communicating with the extracellular matrix, as well as in cellular signaling and can greatly impact cellular function by promoting cell division, apoptosis, or other processes; Type of CAM
Selectins
CAMs that bind to carbohydrate molecules that project from other cell surfaces (these are the weakest CAM bonds); Expressed on white blood cells and the endothelial cells that line blood vessels; Play important role in host defense
Opsonization
When antibodies mark a pathogen for immediate destruction by other white blood cells
Agglutinating
When an antigen and an antibody clump together into large insoluble protein complexes that can be phagocytized and digested by macrophages
Ion Channels
Proteins that create specific pathways for charged molecules; Used for molecules that are impermeable to the membrane (large, polar, or charged); Three types: Ungated, voltage-gated, and ligand-gated
Facilitated Diffusion
A type of passive transport where the diffusion of molecules down a concentration gradient through a pore in the membrane is created by a transmembrane protein ion channel; Used for molecules that are impermeable to the membrane (large, polar, or charged)
Ungated Channels
Have no gates and are therefore unregulated; I.e. Ungated potassium channels allow potassium transport until potassium has reach equilibrium
Voltage-Gated Channels
The gate is regulated by the membrane potential change near the channel; I.e. Sodium channels that are opened during membrane depolarization
Ligand-Gated Channels
The binding of a specific substance or ligand to the channel causes it to open or close; I.e. GABA binds to chloride channel to open it
Enzyme-Linked Receptors
Membrane receptors that display catalytic activity in response to ligand binding; Three primary protein domains: a membrane-spanning domain, a ligand-binding domain, and a catalytic domain
Membrane-Spanning Domain
In enzyme-linked receptors, this anchors the receptor in the cell membrane
Ligand-Binding Domain
In enzyme-linked receptors, this is stimulated by the appropriate ligand and induces a conformational change that activates the catalytic domain
Second Messenger Cascade
A series of events that transmit signals from the outside of a cell to its interior, triggering physiological changes; Caused by the ligand-biding domain inducing a conformational change that activates the catalytic domain
G Protein-Coupled Receptors (GPCR)
A large family of integral membrane proteins involved in signal transduction; Have seven membrane-spanning α-helices; Receptors found on the extracellular surface of the cell; Use a heterotrimeric G protein to transmit signals to an effector in the cell
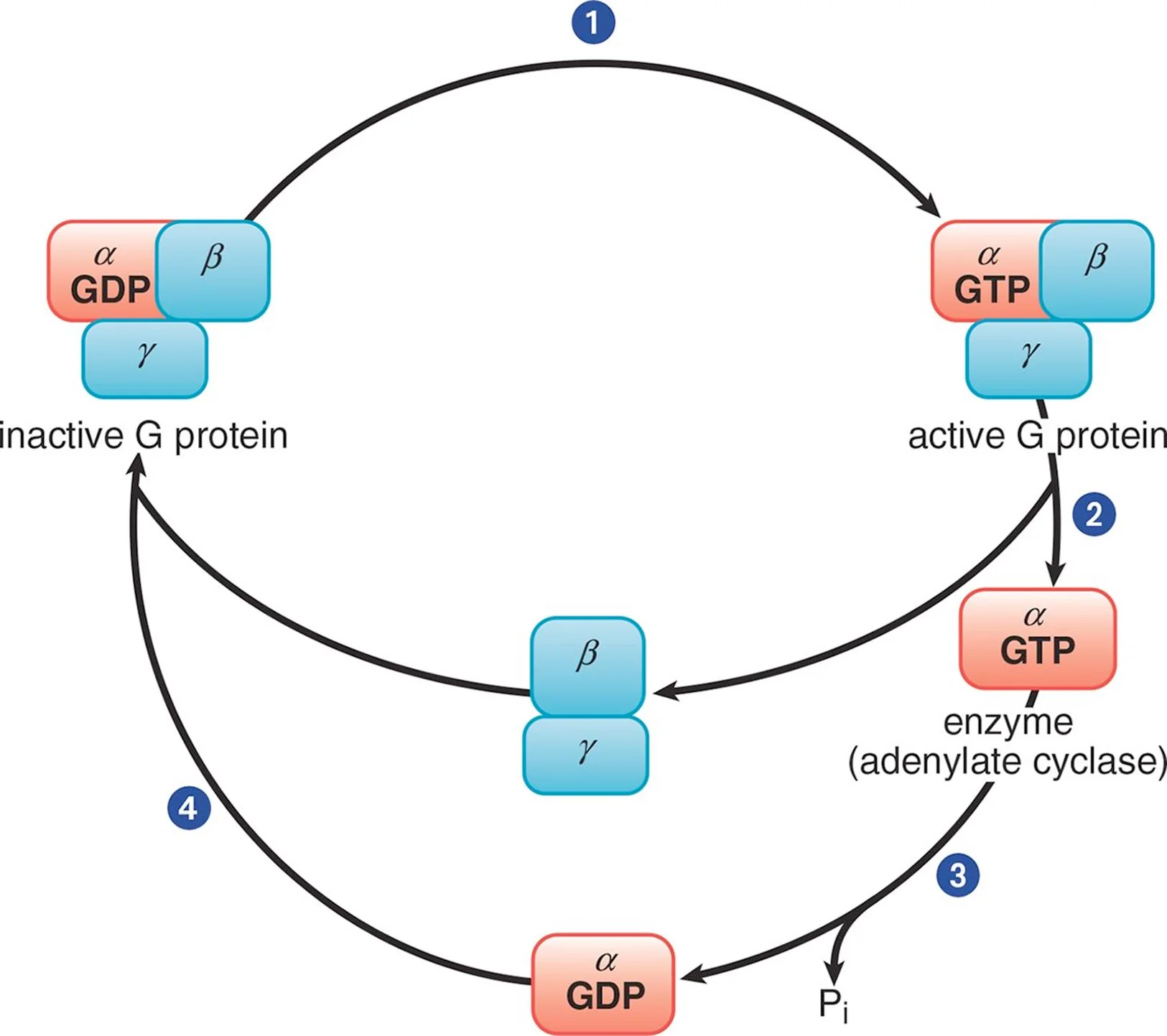
(Heterotrimeric) G Protein
Used by GPCRs to transmit signals to an effector in the cell; Named for their intracellular link to guanine nucleotides (GDP and GTP)
The binding of a ligand increases the affinity of the receptor for the G protein. The binding of the G protein represents a switch to the active state and affects the intracellular signaling pathway.
Three Types: Gs, Gi, and Gq
Gs
Stimulates adenylate cyclase, with increases levels of cAMP in the cell
Gi
Inhibits adenylate cyclase, which decreases levels of cAMP in the cell
Gq
Activates phospholipase C, which cleaves a phospholipid from the membrane to form PIP2. PIP2 is then cleaved into DAG and IP3; IP3 can open calcium channels in the ER, increasing calcium levels in the cell.
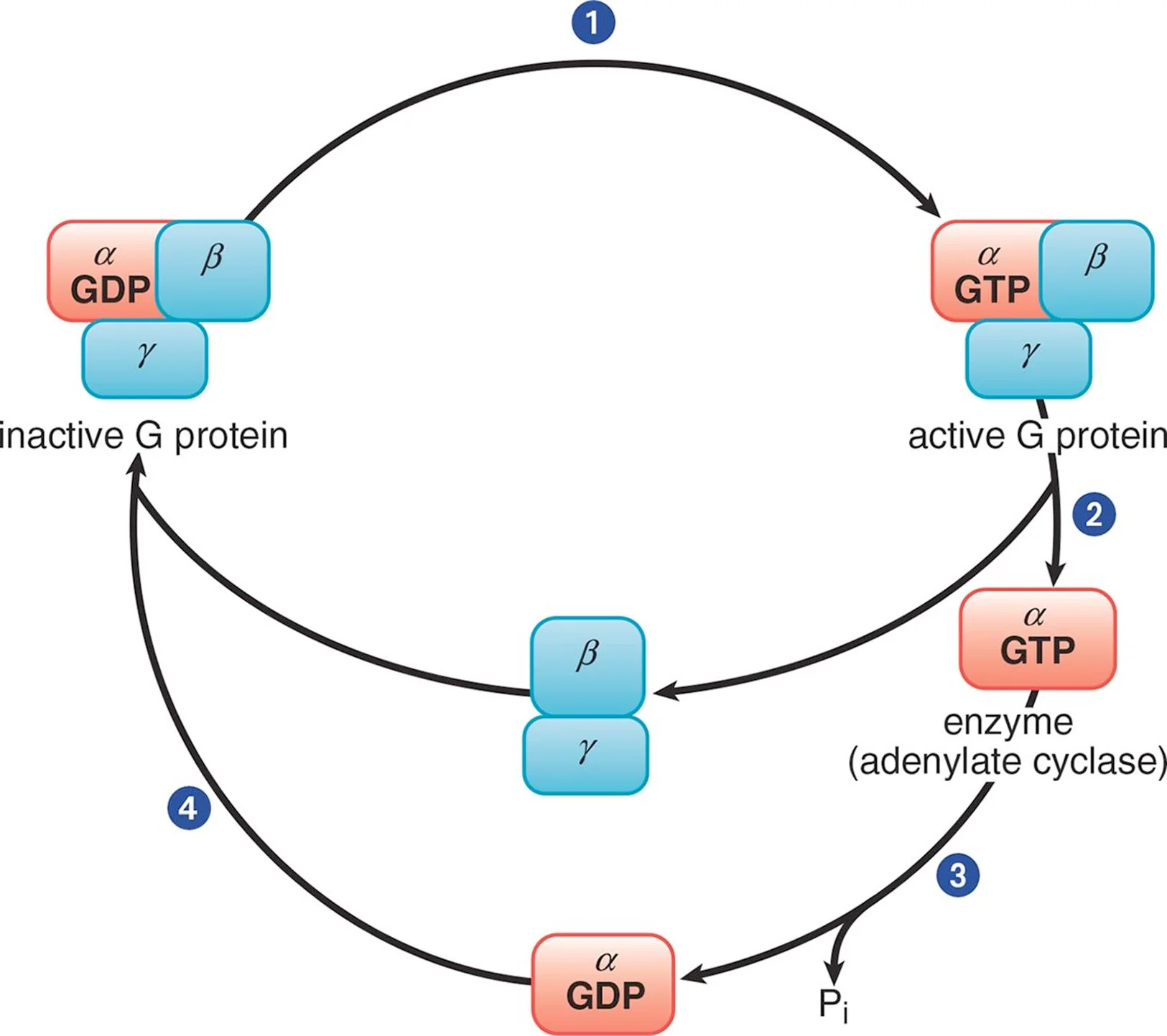
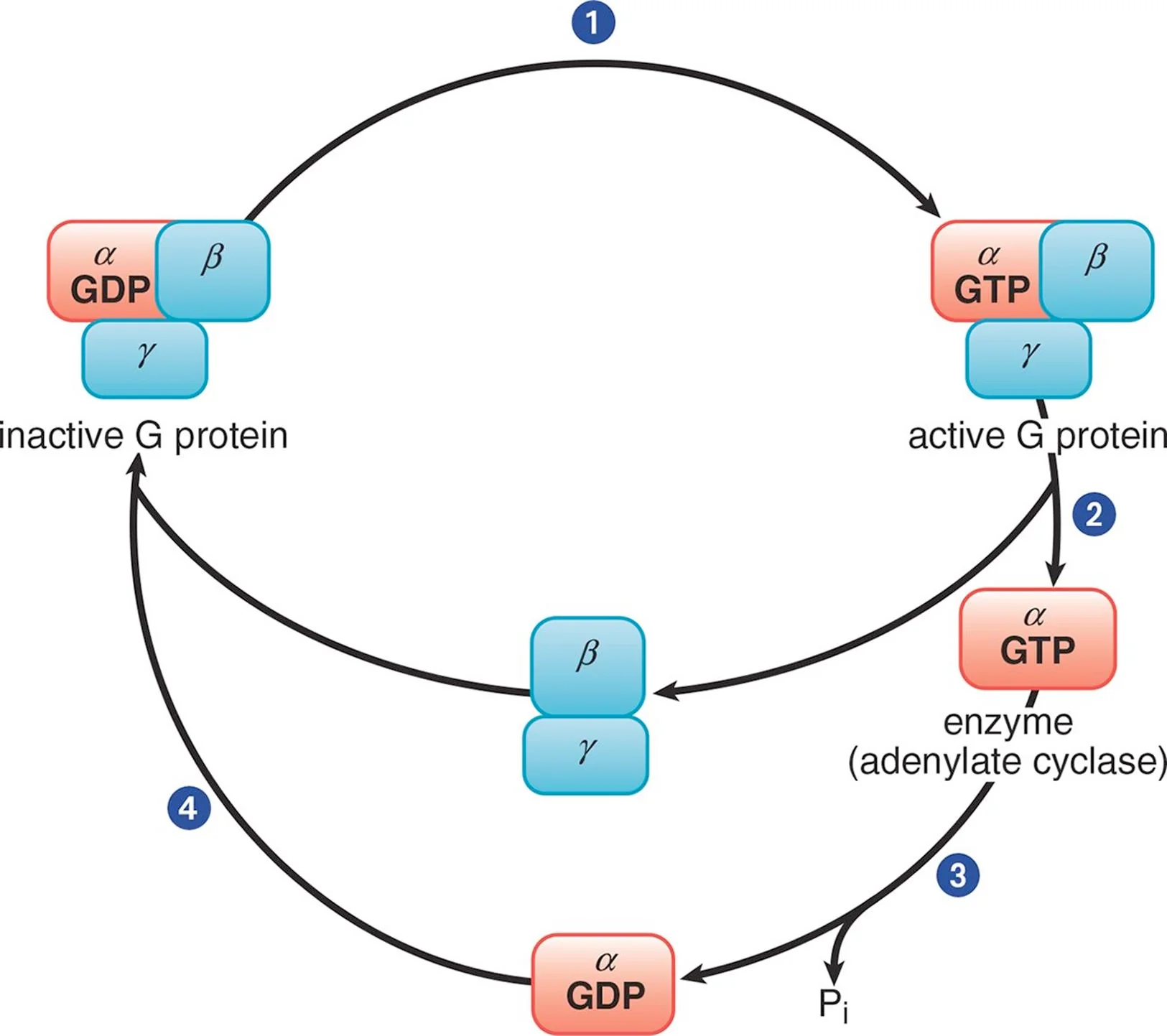
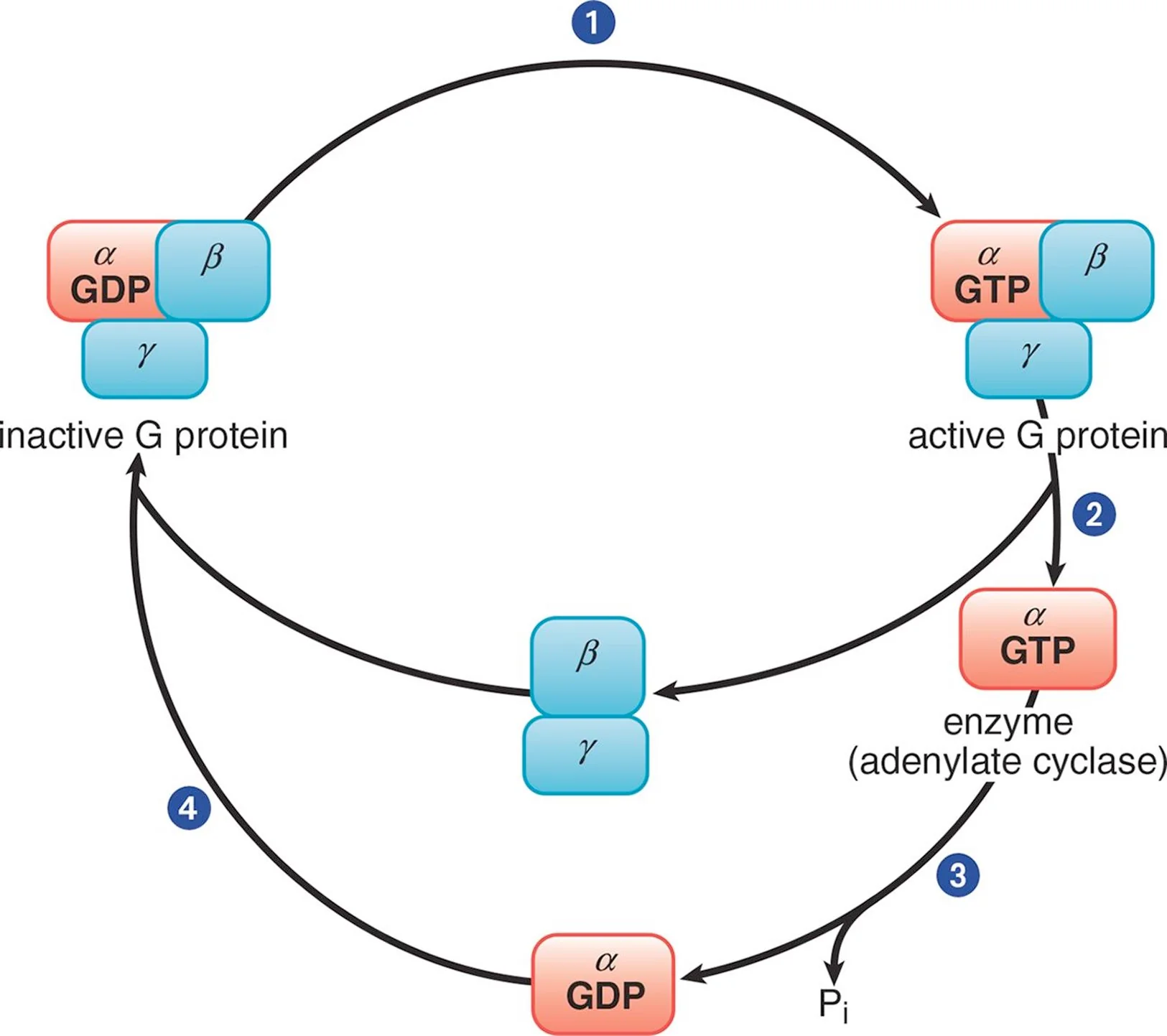
Trimeric G Protein Cycle
In its inactive form, the α subunit binds GDP and is in a complex with the β and γ subunits. When a ligand binds to the GPCR, the receptor becomes activated and, in turn, engages the corresponding G protein (Step 1). Once GDP is replaced with GTP, the α subunit is able to dissociate from the β and γ subunits (Step 2). The activated α subunit alters the activity of adenylate cyclase. If the α subunit is αs, then the enzyme is activated; if the α subunit is αi, then the enzyme is inhibited. Once GTP on the activated α subunit is dephosphorylated to GDP (Step 3), the α subunit will rebind to the β and γ subunits (Step 4), rendering the G protein inactive
Electrophoresis
Works by subjecting compounds to an electric field, which moves them according to their net charge and size. Negatively charged compounds will migrate toward the positively charged anode, and vice versa.
Migration Velocity: v = Ez/f
E = Electric Field Strength
z = Net charge on the molecule
f = Frictional coefficient
Polyacrylamide Gel
The standard medium for protein electrophoresis. Proteins travel through gel in relation to their size and charge, with smaller particles passing through easily while large particles get stuck. Slow molecules: Large and electrically neutral
Polyacrylamide Gel Electrophoresis (PAGE)
Method for analyzing proteins by mass-to-charge and mass-to-size ratios; Functional native protein can be recovered from the gel after electrophoresis (before staining); Useful to compare the molecular size or charge of proteins known to be similar in size from other analytic methods
SDS-PAGE
Separates proteins on the basis of relative molecular mass alone; SDS is a detergent that disrupts all noncovalent interactions and binds to proteins to create large chains with net negative charges, thereby neutralizing the protein’s original charge and denaturing the protein; As proteins move through gel, only affected by electric field strength and friction (depends on mass)
Isoelectric Focusing
Exploits the acidic and basic properties of amino acids by separating on the basis of pI; Protein mixture placed in a gel with a pH gradient (acidic at positive anode and basic at negative cathode) and positively-charged proteins will migrate towards cathode, and vice versa. As the protein reaches the portion of the gel where the pH is equal to the protein’s pI, the protein takes on a neutral charge and will stop moving.
Edman Degradation
Uses cleavage to sequence proteins of up to 50 to 70 amino acids; Selectively and sequentially removes the N-terminal amino acid of the protein, which can be analyzed via mass spectroscopy
Bradford Protein Assay
Mixes a protein in solution with Coomassie Brilliant Blue dye. The dye gives up protons upon binding to amino acid groups, turning blue in the process. Ionic attractions between the dye and the protein then stabilize this blue form of the dye; thus, increased protein concentrations correspond to a larger concentration of blue dye in solution; Very important that only one protein is present
Aldoses
Carbohydrates that contain an aldehyde group as their most oxidized functional group
Ketoses
Carbohydrates that contain a ketone group as their most oxidized functional group
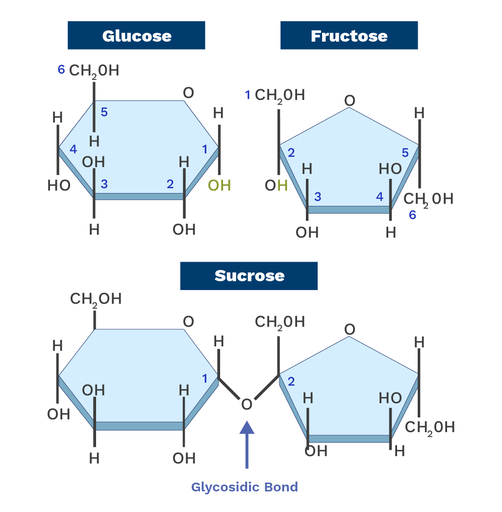
Glycosidic Linkages
A covalent bond that joins a carbohydrate molecule to another group, such as another carbohydrate or an alcohol; Sugars acting as substituents via this linkage are called glycosyl residues
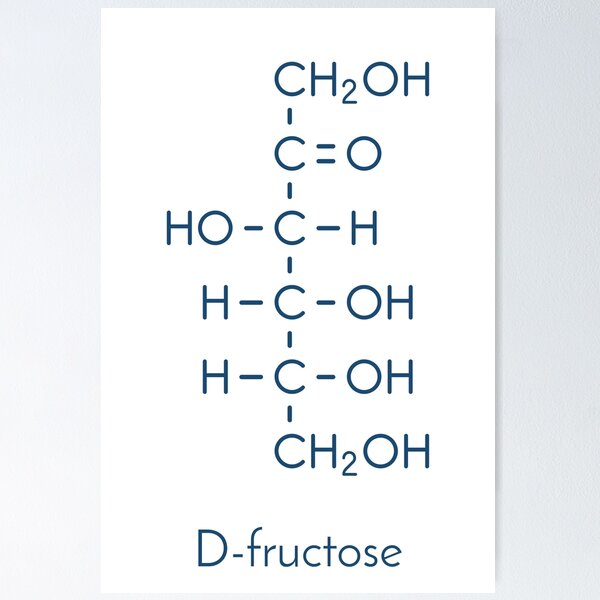
D-Fructose
D-Glucose

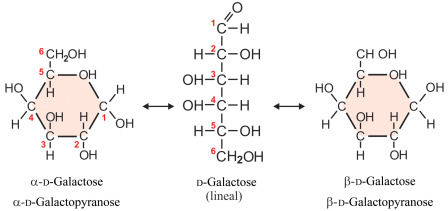
D-Galactose
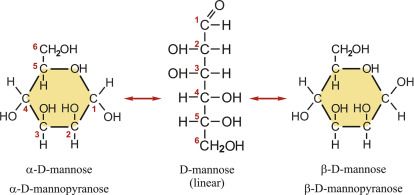
D-Mannose
Number of Possible Stereoisomers
Number of stereoisomers with a common backbone = 2^n
n = Number of chiral carbons in the molecule
L- vs. D-Sugars (Fischer Projection)
All D-sugars have the hydroxide of their highest-numbered chiral center on the right, and all L-sugars have that hydroxide on the left (enantiomers)
Epimers
A type of diastereomer that differs in configuration at exactly one chiral center
Hemiacetals
Cyclic molecules formed from aldoses
Hemiketals
Cyclic molecules formed from ketoses
Anomeric Carbon
A carbon atom in a cyclic sugar that was once the carbonyl carbon in the open-chain form of the sugar
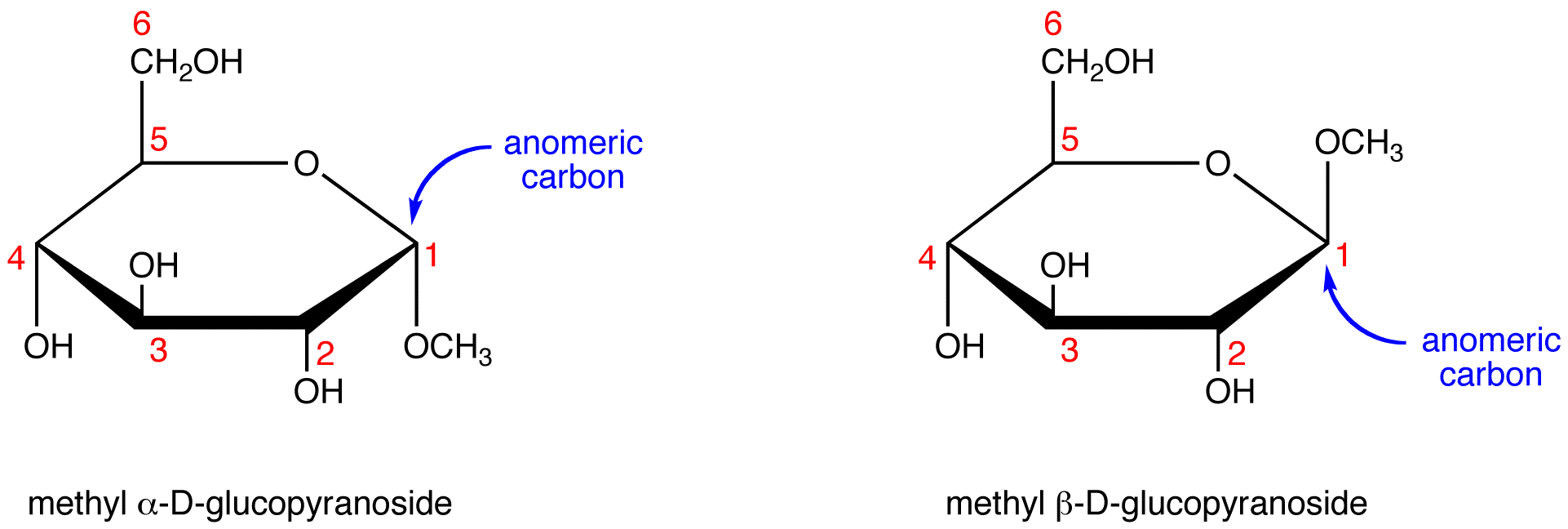
Anomers
Two sugars in ring formation that differ at the anomeric carbon
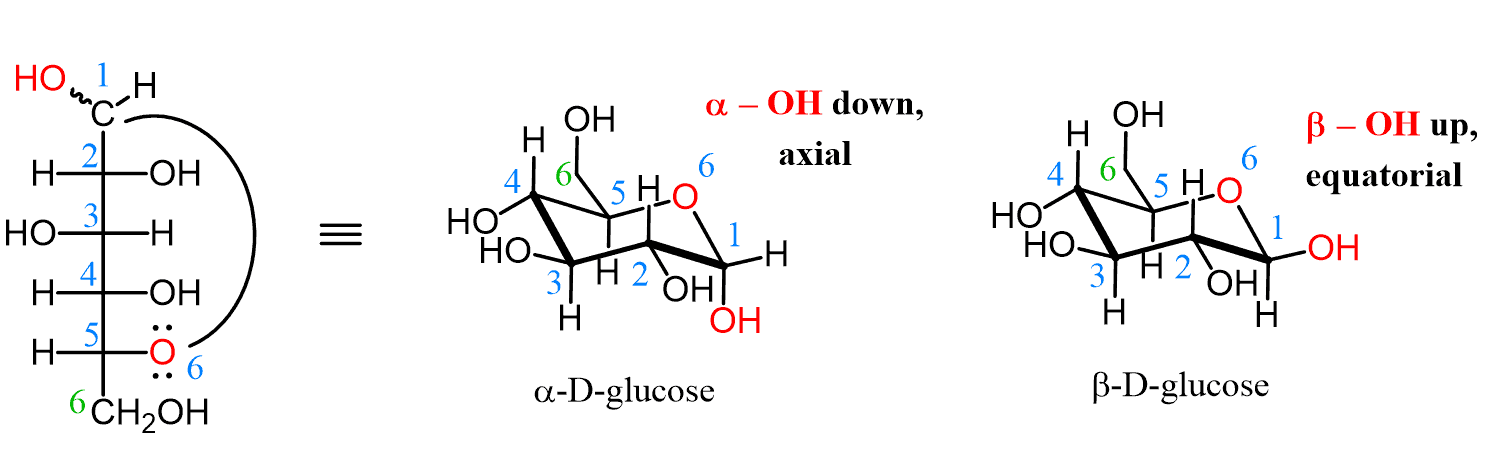
α-anomer
In glucose, this has the -OH group of C-1 trans to the -CH2OH substituent (axial and down)
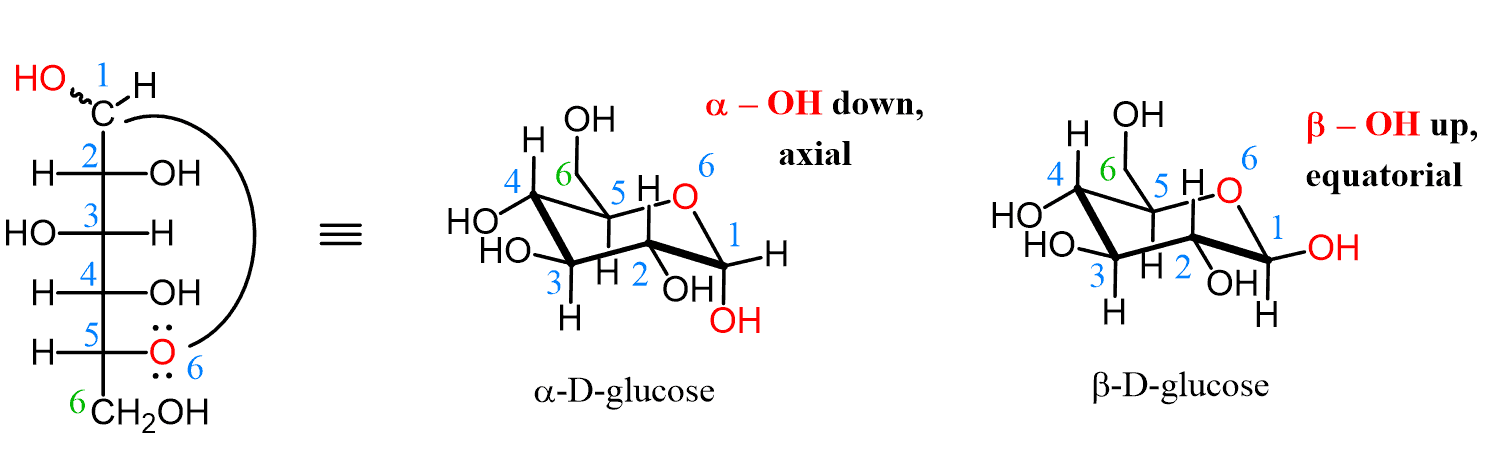
β-anomer
In glucose, this as the -OH group of C-1 cis to the -CH2OH substituent (equatorial and up)
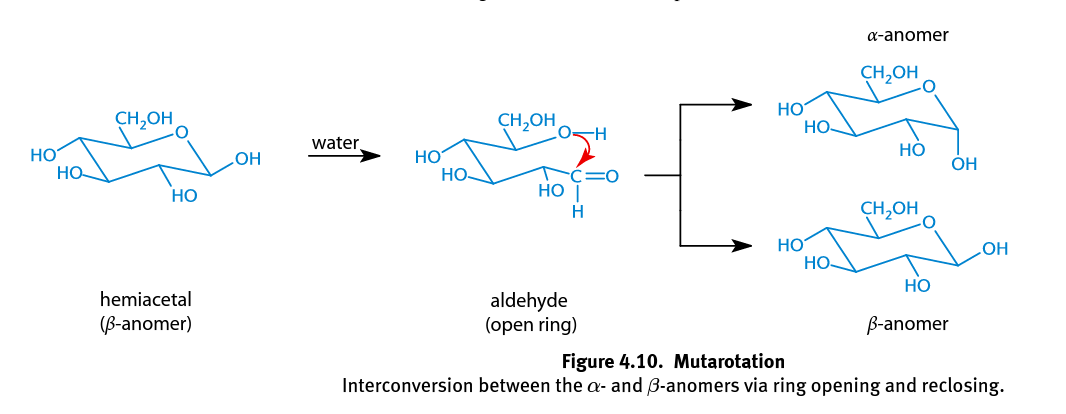
Mutarotation
In hemiacetal rings the single bond between C-1 and C-2 can rotate freely, and either the α- or β-anomer can spontaneously form; Occurs more rapidly when catalyzed with an acid or base; In solution, the α-anomeric configuration is less favored because the hydroxyl group of the anomeric carbon is axial, adding to the steric strain of the molecule
Aldonic Acids
Oxidized aldoses; Often oxidized by the human body in order to yield energy (reducing agents)
Reducing Sugar
Any monosaccharide with a hemiacetal ring
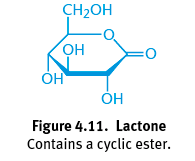
Lactone
Product of the oxidation of an aldose in ring form; A cyclic ester with a carbonyl group persisting on the anomeric carbon; Essential role in human body
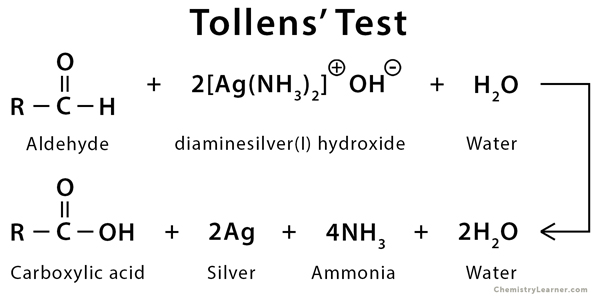
Tollens’ Reagent
Produces a silvery mirror when the aldehyde group is readily available; Ketoses also test positive due to tautomerization under basic conditions to form a carboxylic acid
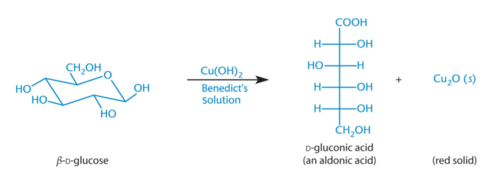
Benedicts’ Reagent
The aldehyde group of an aldose is readily oxidized, creating red Cu2O; Ketoses also test positive due to tautomerization under basic conditions to form a carboxylic acid
Tautomerization
The rearrangement of bonds in a compound, usually by moving a hydrogen and forming a double bond
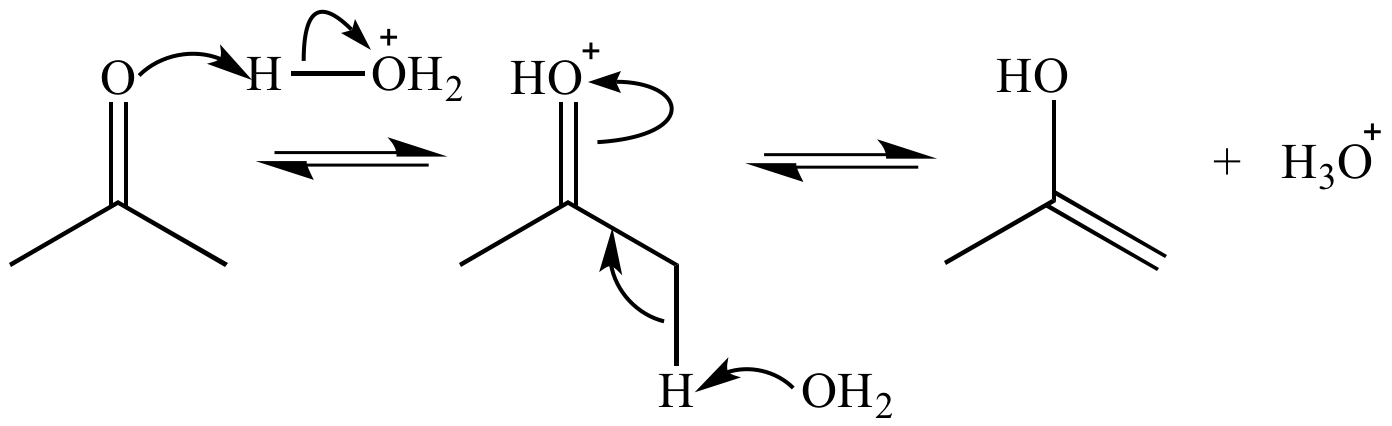
Enol
A compound with a double bond and an alcohol group
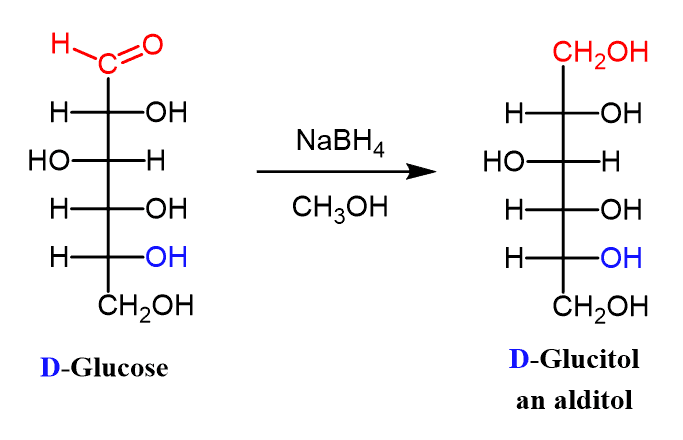
Alditol
When the aldehyde group of an aldose is reduced to an alcohol
Deoxy Sugar
Contains a hydrogen that replaces a hydroxyl group on a sugar; Ex. D-2-deoxyribose
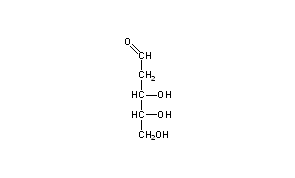
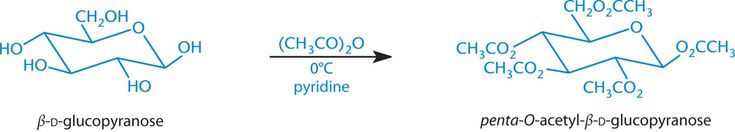
Esterification of Carbohydrates
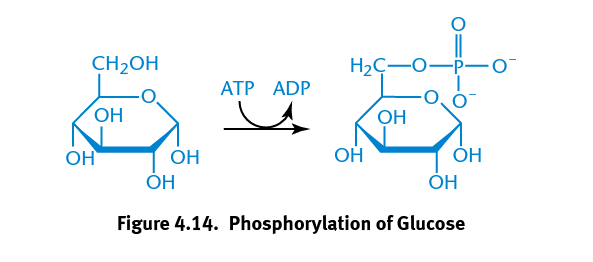
Hexokinase
Catalyzes the phosphorylation of glucose important to metabolic reaction of glycolysis in which a phosphate group is transferred from ATP to glucose
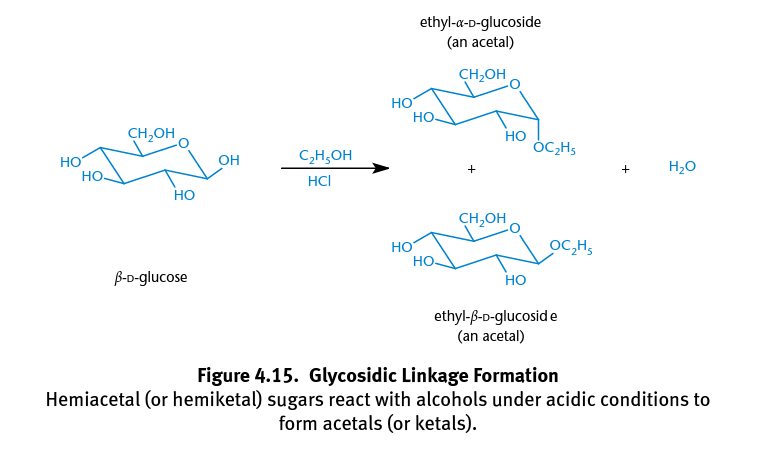
Acetals
Formed when hemiacetals react with alcohols; The anomeric hydroxyl group is transformed into an alkoxy group, creating α- and β-acetals; Creates glycosides formed by glycosidic bonds
Furanosides
Glycosides derived from furanose (5-member carb) rings
Pyranosides
Glycosides derived from pyranose (six-member carb) rings
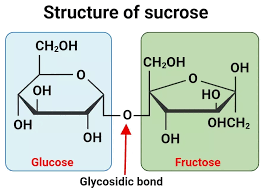
Sucrose
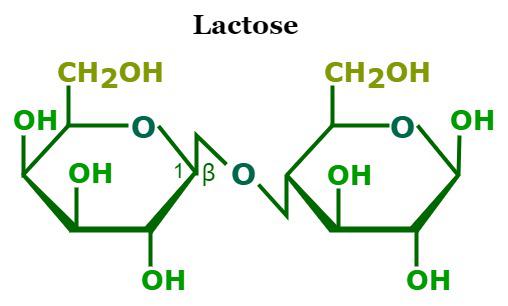
Lactose
Glucose + Galactose
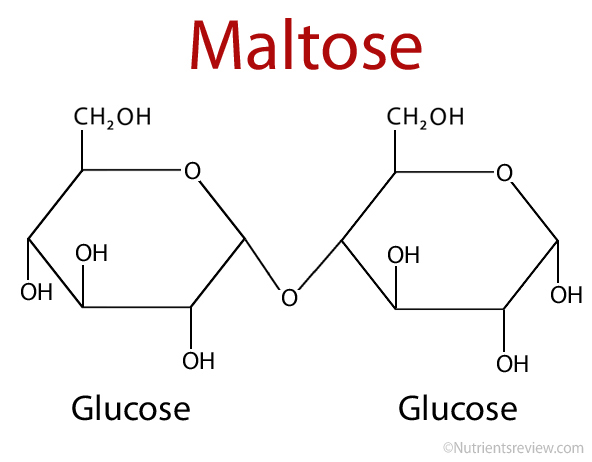
Maltose
Glucose + glucose
Homopolysaccharide
A polysaccharide composed entirely of glucose (or any other monosaccharide)
Heteropolysaccharide
A polymer made up of more than one type of monosaccharide
Cellulose
The main structural component of plants; Homopolysaccharide composed of β-D-glucose molecules linked by β-1,4 glycosidic bonds, with hydrogen bonds holding the actual polymer chains together for support
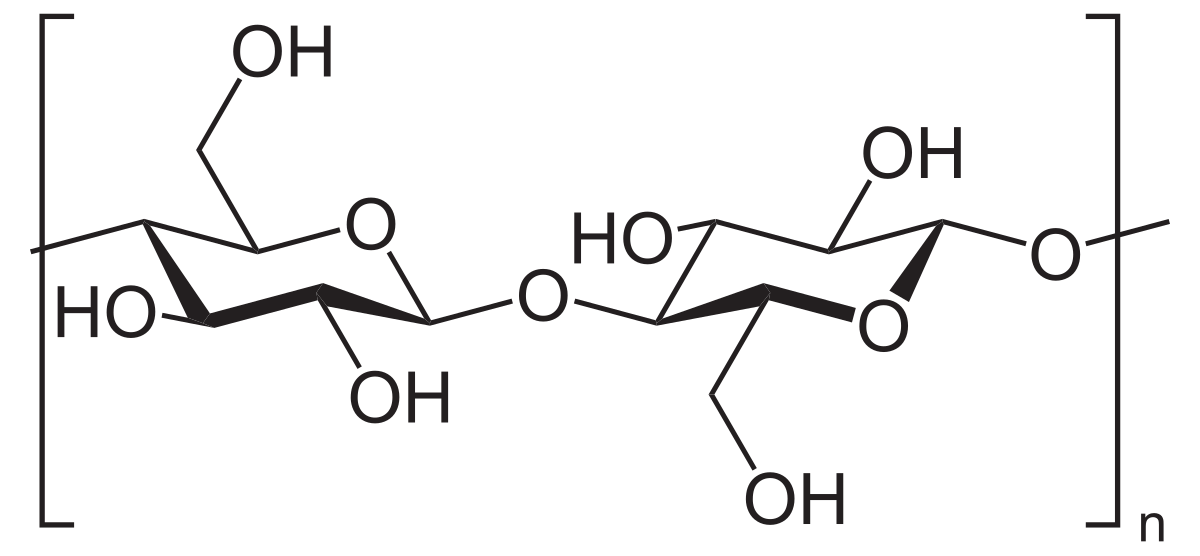
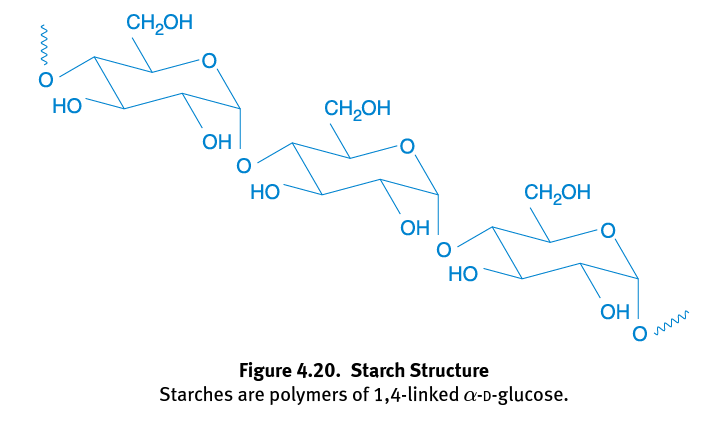
Starches
Polysaccharides that are more digestible by humans because they are linked α-1,4 glycosidic bonds; Can be broken down by enzymes and used as a source of energy
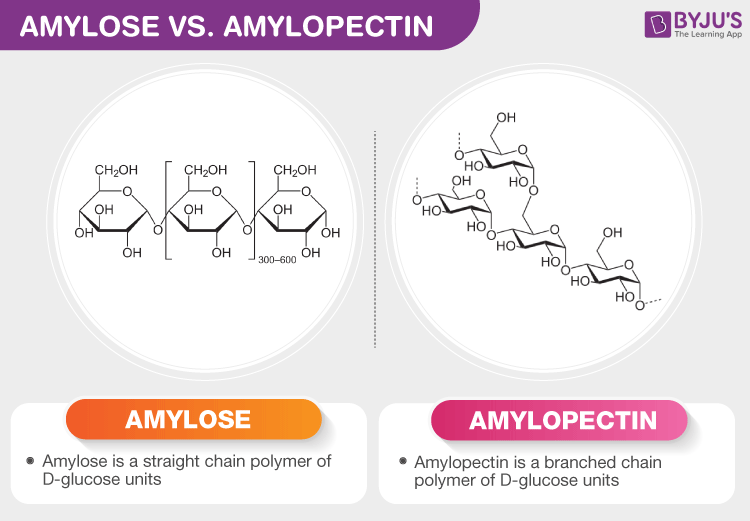
Amylose
A linear glucose polymer linked via α-1,4 glycosidic bonds; How plants predominantly store starch
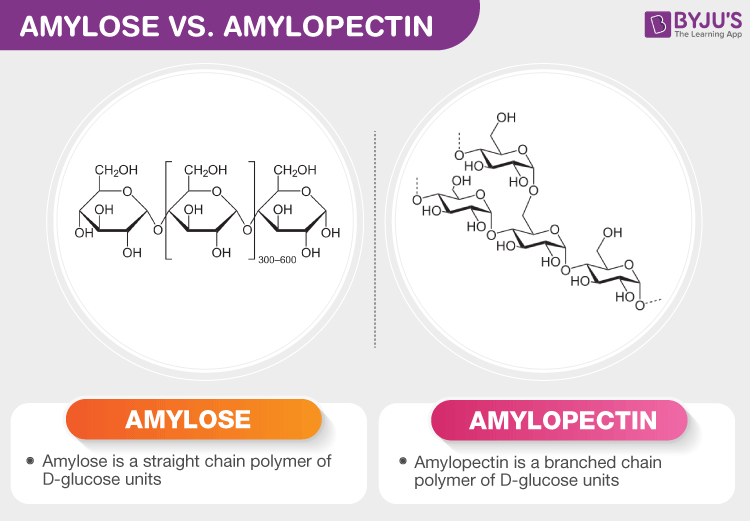
Amylopectin
Type of starch similar to amylose but contains branches via α-1,6 glycosidic bonds