Unit 3: DNA Repair
1/63
Earn XP
Description and Tags
Study guide, in class exercise, definitions and guide
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
64 Terms
Describe the two types of spontaneous DNA damage
The two types of spontaneous DNA damage are depurination and deamination. They are classified as spontaneous because the damage is naturally occurring and random and is independent from DNA replication.
Depurination- Depurination can remove guanine or adenine from DNA. Depurination does not break the DNA phosphodiester backbone but instead removes a purine base from a nucleotide.
Deamination - The major type of deamination converts cytosine to an altered DNA base, in this case uracil. Deamination is "the spontaneous loss of an amine group..."
Both deamination and depurination take place on double helical DNA, and neither break the phosphodiester backbone. These are modifications to single bases on one strand that will be fixed by excision repair.
What are the possible consequences of not being able to repair each of the types of spontaneous DNA damage?
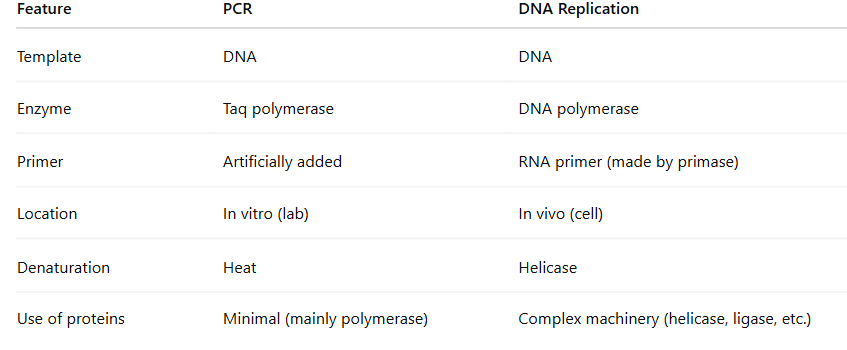
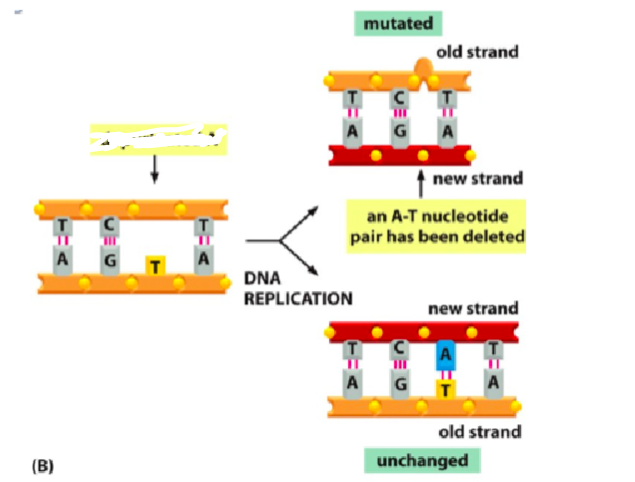
The consequence of spontaneous DNA damage is that it may result in a change of a codon and incorrect amino acid sequences in the protein. This can lead to altered protein function or incorrect expression of RNA. The unrepaired deamination can change the codon and lead to an amino acid substitution or nonsense mutation. The unrepaired depurination will lead to nucleotide deletion and shift how the remaining information for the rest of the protein sequence will be read during translation. This is referred to as a “frame shift” mutation and will impact more than a single amino acid.
Describe the example that was provided of induced DNA damage
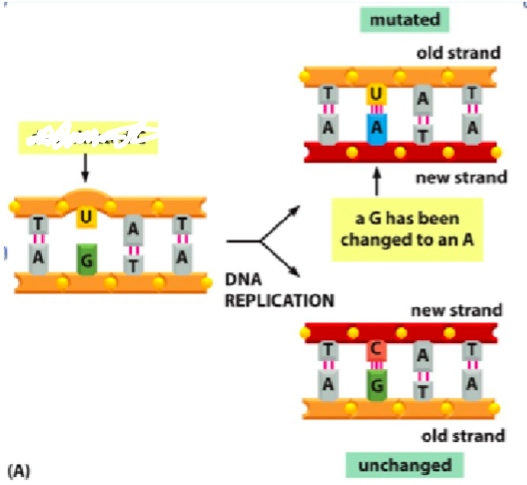
Ultraviolet radiation in sunlight is damaging to DNA in that it can cause the formation of thymine dimers, covalent linkage between two adjacent pyrimidine bases. This can block the passage of the DNA replication polymerase in the cell that contains this damage.
What are the possible consequences of not being able to repair induced DNA damage?
Possible consequences of failing to repair induced DNA damage (i.e. from ultraviolet light exposure) include the formation of pyrimidine dimers, which stall the replication machinery and can cause mutations to accumulate.
What types of DNA damage can be corrected by the excision repair mechanism?
There are three types of excision repair: Mismatch repair, base excision repair (BER) and nucleotide excision repair (NER). Damage that can be corrected by the excision repair mechanism is limited to single strand mutations. This includes incorrect base-pairs (which create a bulge in the sugar-phosphate backbone and are corrected through the mismatch repair pathway), spontaneous DNA damage (random mutation via depurination or deamination…these modifications are made to the bases and do not impact the sugar phosphate backbone) usually repaired by BER, and induced DNA damage (by UV radiation or chemical exposure) which creates bulky lesions that are repaired by NER.
What steps/enzymes with the excision repair process will be the same no matter which type of single strand DNA damage is being repaired?
To tie together the process, below is a brief description of how the process generally works:
1). Recognize and remove: The single stranded damaged DNA is recognized and removed.
These steps involve different enzymes that recognize the specific type of DNA damage and nucleases (nucleases are specific for the type of DNA damage), which cleave the covalent bonds that join the damaged nucleotides to the rest of the DNA strand, leaving a small gap on one strand of the DNA double helix region. After removal of the damage, there is a clean template region and the remaining steps can use the same enzymes to synthesize and ligate.
2). Copy : A repair DNA polymerase bind the 3’-OH end of the cut DNA strand. It then fills in the gap by making a complimentary copy of the information stored on the undamaged strand. Although different from the type of DNA polymerase that replicates the leading strand, repair DNA polymerases synthesize DNA strands in the same way. For example, they elongate chains in the 5’-to-3’ direction and have the same type of proofreading activity to ensure that the template strand is copied accurately. In many cells, this is the same enzyme that fills in the gap left after the RNA primers are removed during the normal DNA lagging strand replication process.
3). Seal: When the repair DNA polymerase has filled in the gap, a break remains in the sugar-phosphate backbone of the repaired strand. This nick in the helix is sealed by DNA ligase, the same enzyme that joins the Okazaki fragments during replication of the lagging DNA strand.
Is the repair polymerase used during excision repair the same enzyme used during DNA replication? If different, what are the similarities/difference between the excision repair and replication polymerases?
The repair polymerase used during excision is not the same enzyme used during DNA replication on the leading strand or to make the Okazaki fragments. It is the same enzyme used to fill in the sequences missing once the RNA primer has been removed.
DNA repair polymerase and DNA replication polymerase both synthesize the strands in the same way. They both elongate chains in the 5'-to-3' direction, require a 3’OH to initiate the synthesis and have the same type of proofreading activity.
What are two mechanisms of DNA double strand break repair? What are possible impacts of using each of these types of double strand break repair?
Nonhomologous End-Joining- After a double strand break, when a homologous chromosome is not available, the broken ends are digested back by a nuclease to create blunt ends that can be joined by DNA ligase. The net result is a repaired DNA molecule with nucleotide deletions at the repair site. This is an “emergency repair mechanism” because it risks losing genetic information if the damage was in a coding region or a region essential for regulating splicing, transcription, etc..
Homologous Recombination- If a double strand break occurs shortly after DNA replication but before the chromatids separate in m-phase, the sister chromatid can be used to accurately repair the break with no loss of genetic information. A nuclease digests the 5’ ends, then one strand invades the neighboring double helix by finding a complementary base pair sequence. Repair polymerase synthesizes the missing DNA in the 5’ 3’ direction. The broken double helix is reformed, and DNA synthesis continues using the repaired strand as a template for the damaged complementary strands. DNA ligation restores the original double helix without loss of genetic information.
How are these two mechanisms of DNA repair different than excision repair?
1. The main difference between the two mechanisms of DNA double strand break repair (nonhomologous end-joining and homologous end-joining) and excision repair is that excision repair only involves the repair of one strand of DNA damage while the other two mechanisms involve the repair of two strands of DNA damage
2. The excision repair functions and the homologous end joining mechanism are both maintaining the genetic information. This sharply contrasts the requirements for nonhomologous end-joining repair of DNA double strand damage in that nonhomologous end-joining does not maintain all the genetic information.
What are possible impacts of using each of these types of double strand break repair?
In nonhomologous end-joining you will have a loss of some of the nucleotides. The nuclease repairs the damage by evening out the break on both strands, those nucleotides that were removed are lost.
In homologous end-joining when one DNA helix is broken in a homologous pair, the other DNA helix can act as a template to repair the damage. In this method, there is no loss of genetic information. In most cases of homologous recombination, the sister chromatid is used as the template and the exact same genetic information is copied. However, infrequently the homologous chromosome is used as the template and they may not be the exact same nucleotide sequences that were lost from the break on the first double helix (the homologous chromosome may have a different allele of the gene).
What are possible benefits to mutations?
Mutations can increase genetic diversity, which is essential for evolution and adaptation.
They can lead to new or improved protein functions (e.g., antibiotic resistance in bacteria).
Some mutations confer advantageous traits, such as resistance to disease or improved metabolism.
What are possible drawbacks to mutations?
Mutations can disrupt gene function and lead to diseases like cancer or genetic disorders (e.g., cystic fibrosis).
They can result in nonfunctional or toxic proteins.
Harmful mutations can be passed on to offspring, affecting future generations.
What are possible sources of DNA damage?
Endogenous: Reactive oxygen species, replication errors, spontaneous deamination.
Exogenous: UV radiation, ionizing radiation, chemical mutagens, environmental toxins
When there is a mismatch, how does the cell know which strand of DNA has the wrong nucleotide?
In prokaryotes, the parental strand is methylated (e.g., at GATC sequences), and the newly synthesized strand is unmethylated for a short time—this allows mismatch repair proteins to identify and correct the new strand.
In eukaryotes, the mechanism is less understood but likely involves recognition of nicks in the newly synthesized strand (before ligation is complete).
If deamination occurs, what are two ways the cell can tell it is a mutation?
Uracil in DNA (from cytosine deamination) is not normal—DNA repair enzymes like uracil-DNA glycosylase recognize and remove it.
Thymine from 5-methylcytosine deamination is harder to detect, but the mismatch with guanine can be recognized by mismatch repair machinery.
Since nucleotides are lost in non-homologous end joining, would it be better to not repair the double-strand break?
No. Unrepaired double-strand breaks are extremely dangerous and can lead to chromosomal fragmentation, cell death, or cancer.
Even though NHEJ is error-prone, it is better than leaving the break unrepaired.
When do you use non-homologous end joining over homologous end joining and vice versa?
NHEJ: Used throughout the cell cycle but especially in G1, when a homologous template is not available.
HR: Used in S and G2 phases, when a sister chromatid is available as a template for accurate repair.
Do all mutations result in coding DNA damage?
No.
Many mutations occur in non-coding regions (introns, intergenic regions).
Some are silent mutations that do not change the amino acid.
Others affect regulatory elements or splicing, which may or may not have functional
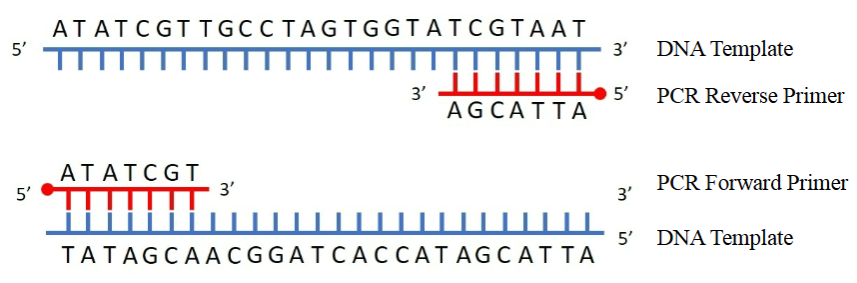
What are the similarities and differences between PCR and DNA replication
What primers would we use to amplify the sequence starting at ATG and ending with GCTA?
5’AAAACTGATGGCACTCTGCAT…………………….TCTGAGATCCCAAGGTTGCTATTCCATGC3’
Given sequence:
5′ AAAAACTGATGGCACTCTGCAT…………………….TCTGAGATCCCAAGGTTGCTATTCCATGC 3′
We want:
Forward primer starting at ATG = ATGGCACTCTGCAT (5′ to 3′)
Reverse primer = reverse complement of the end at GCTA (GCTA = AGCA in template, reverse complement is GCATGGAATAGCAACCTTGGGATCTCAGA)
So:
Forward primer: 5′ ATGGCACTCTGCAT 3′
Reverse primer: 5′ GCATGGAATAGCAACCTTGGGATCTCAGA 3′
Describe the process of automated DNA sequencing
DNA is denatured to single strands.
A primer binds to the template strand.
DNA polymerase adds nucleotides from a mix of normal dNTPs and fluorescently labeled ddNTPs.
When a ddNTP is incorporated, chain extension stops.
The result is a mixture of fragments of different lengths, each ending in a ddNTP.
Fragments are separated by capillary electrophoresis.
A laser detects the fluorescent label on each terminating base.
A computer reads the sequence based on the colors and sizes of the fragments.
Is there sequence selectivity for where a transposon will insert?
Some transposons show weak sequence preference (e.g., for palindromic or AT-rich sequences), but many insert somewhat randomly.
Target site duplication often occurs regardless of sequence specificity.
Which transposon mechanism is the only one that will not result in a net increase in the number of transposons in the genome?
"Cut-and-paste" DNA transposition
The transposon is excised and moved to a new location—no copy is left behind
Which transposon mechanism requires an RNA intermediate
Retrotransposition (e.g., LINEs, SINEs, retroviruses)
Involves transcription into RNA, reverse transcription into DNA, and integration into a new site.
What is this diagram
Depurination
What is this diagram
Deamination
What is this diagram
Induced damage
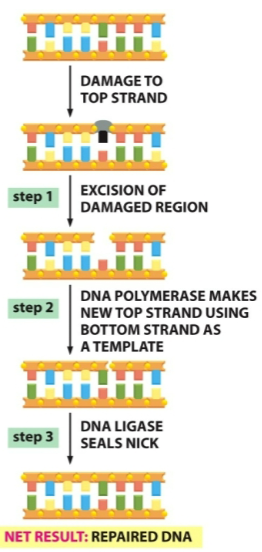
What is this diagram
Excision repair
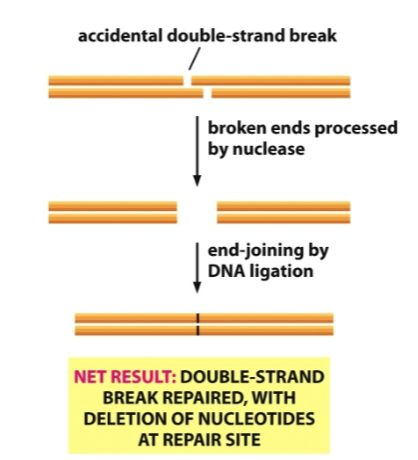
What is this diagram
Nonhomologous end joining
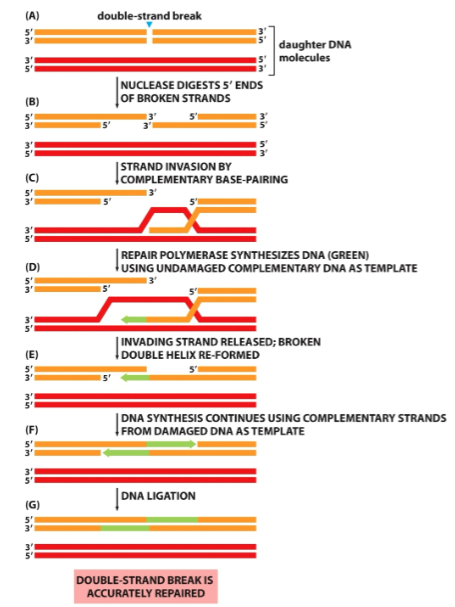
What is this diagram
Homologous recombination
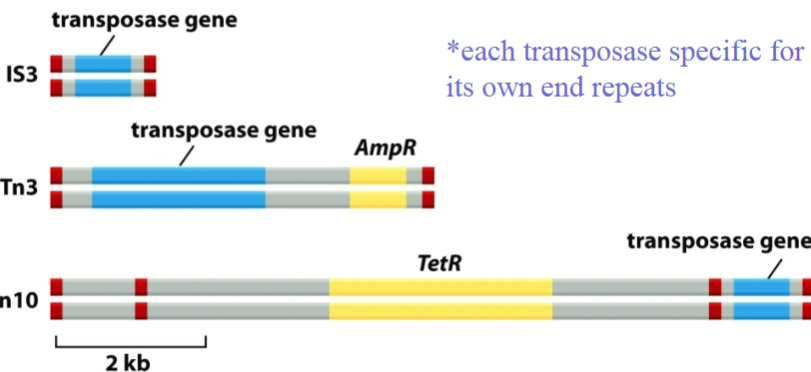
What is this a diagram of and what are the 2 required components
Mobile genetic elements are another source of DNA variation
Transposons have transposase and nucleotide repeats
What are the three types of transposition
Cut and paste (DNA), replicative transposition (DNA), and retrotransposition (RNA)
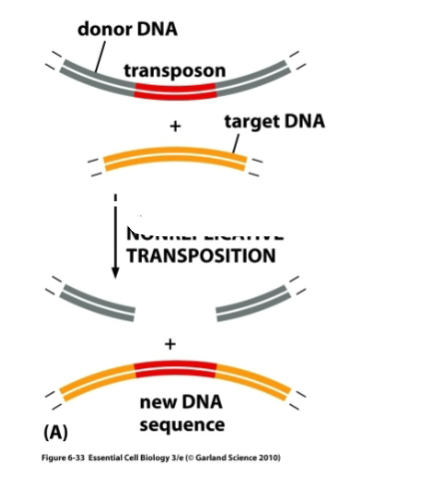
What is this a diagram of
Cut and paste transposition
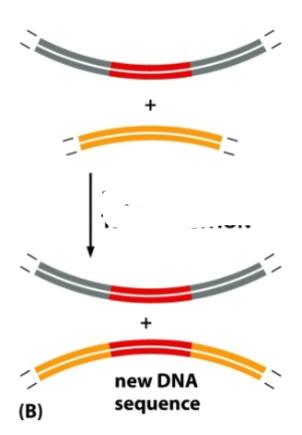
What is this a diagram of
Replicative transposition
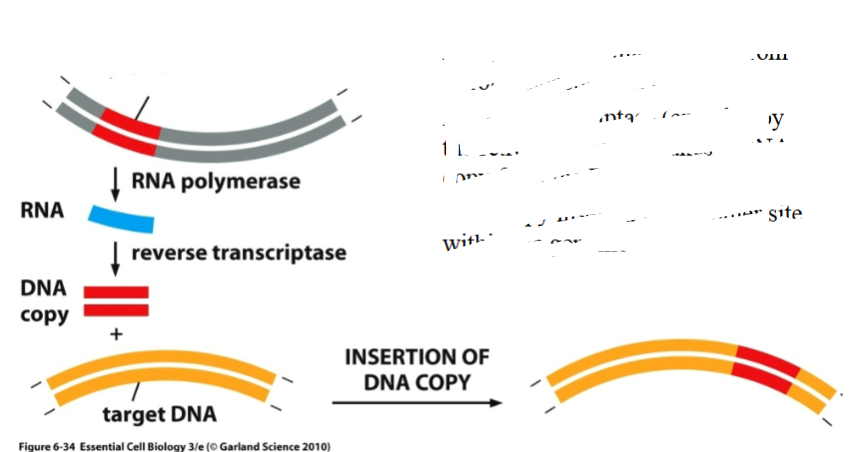
What is this a diagram of
Retrotransposition
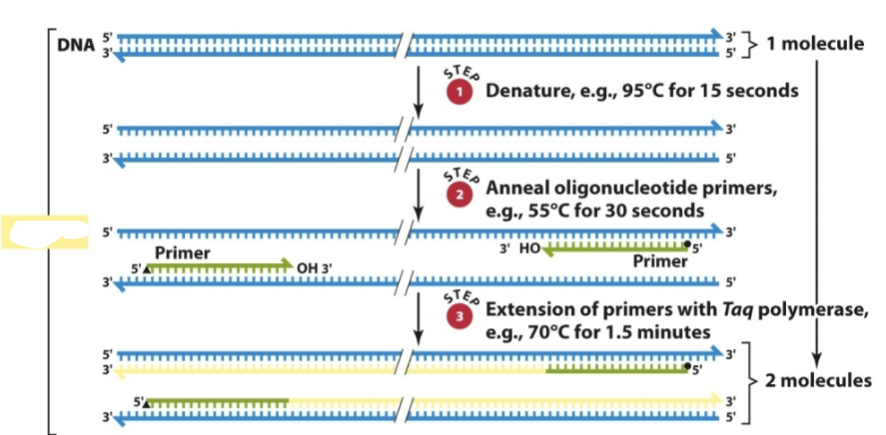
What are the 3 repeated steps to PCR
1) Denaturation of genomic DNA (92-95oC)
2) Annealing of denatured DNA to oligonucleotide primers (50-60oC) Synthetic nucleotides complementary to known flanking sequences are
used to prime enzymatic amplification of the sequence of interest.
3) Extension - Replication of the DNA segment between primer
binding sites (70-72oC).
• Amplification occurs exponentially; each cycle doubles the number
of molecules of the sequence of interest.
• Thermocycler (PCR machine) facilitates rapid temperature changes
of samples.
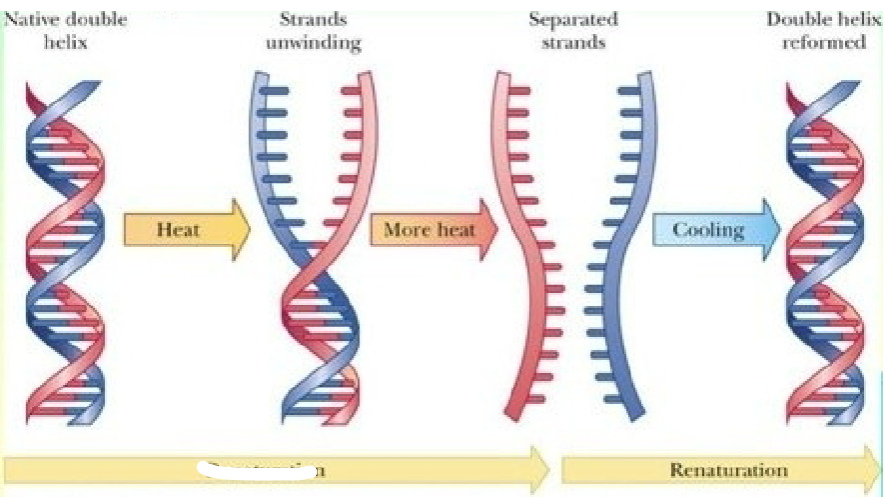
What is this a diagram of
Denaturation
What is this a diagram of
Annealing
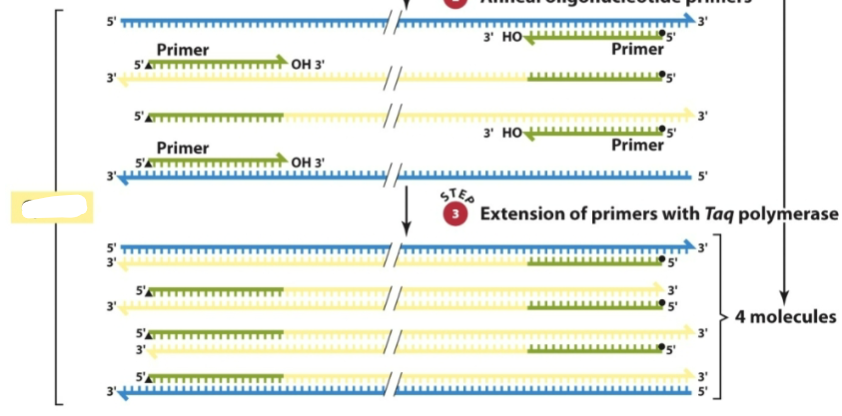
What is this a diagram of
PCR cycle 1
What is this a diagram of
PCR cycle 2
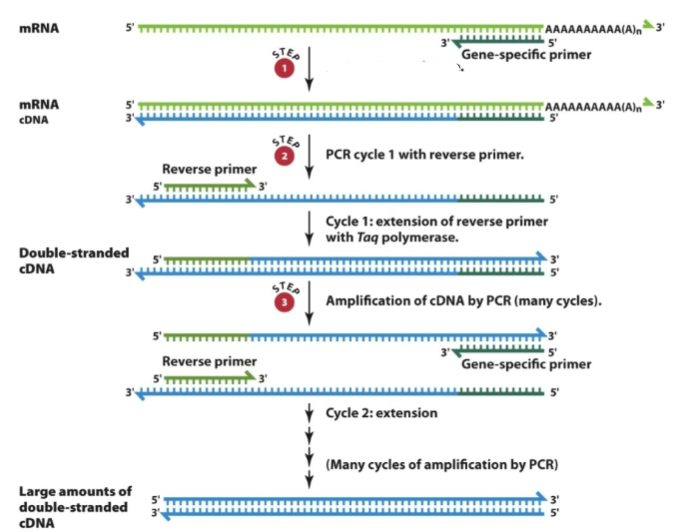
What is this a diagram of
RT-PCR can also mean
Reverse Transcriptase-PCR (RT-PCR)
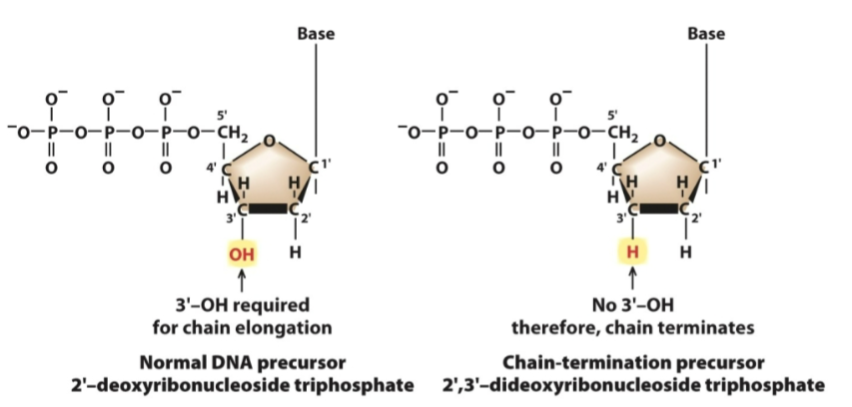
What is this a diagram of
2',3'-Dideoxyribonucleoside Triphosphates - used in DNA sequencing
These are the chain-terminating nucleotides used in Sanger sequencing. The slide compares them to normal DNA nucleotides (dNTPs), which allow DNA to be extended.

What is DNA sequencing
A population of DNA fragments is generated using
DNA polymerase and nucleotide bases.
– One end is common to all fragments (the 5’ end of the sequencing primer).
– The other end terminates at all possible positions (the 3' terminus).
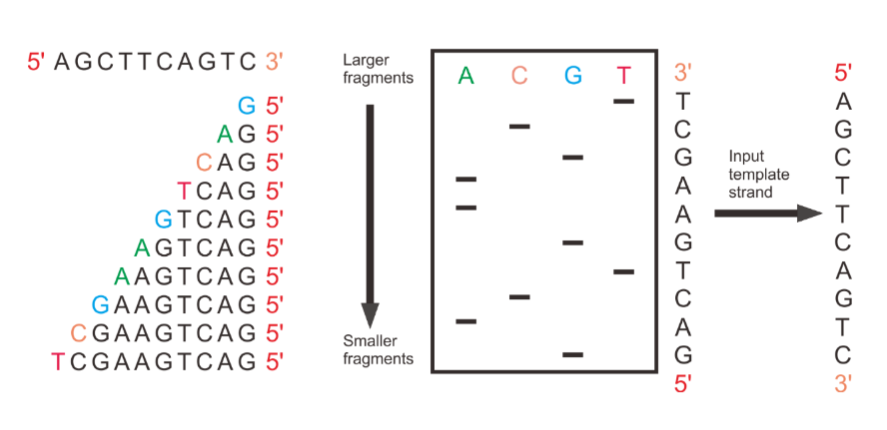
Explain this diagram
The sequencing reactions on the left.
A sequencing gel in the center.
The input template strand on the right.
The final sequence readout at the top left.
Describe Automated DNA Sequencing
Fluorescent dyes are used for detection of DNA chains instead of radioactive isotopes.
Products of all four chain terminator reactions are
separated through a single gel or capillary tube.Photocells detect fluorescence of the dyes as they pass through the gel or capillary tube.
Output of the photocell is directly transferred to a
computer for analysis.
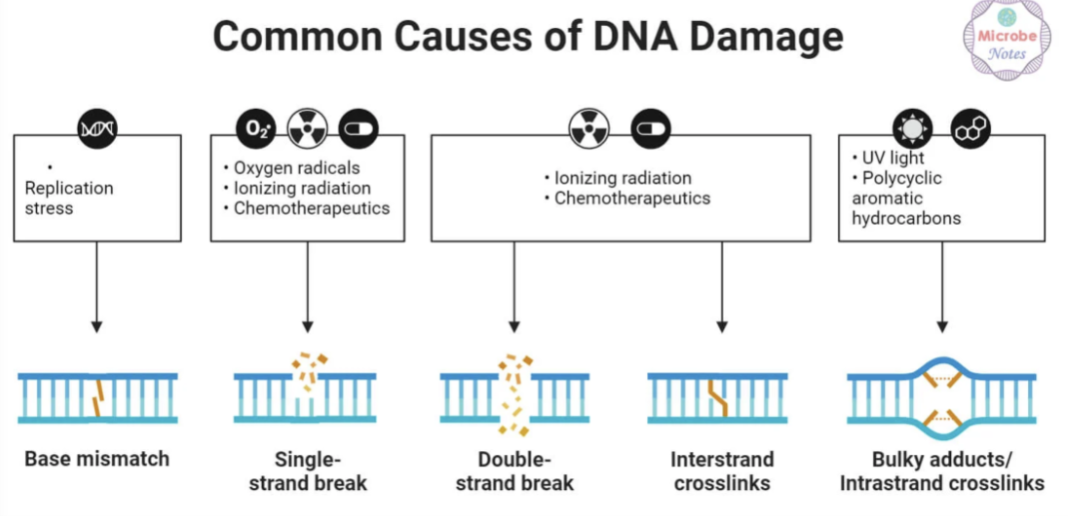
Identify the common causes of DNA damage and the type of damage it produced
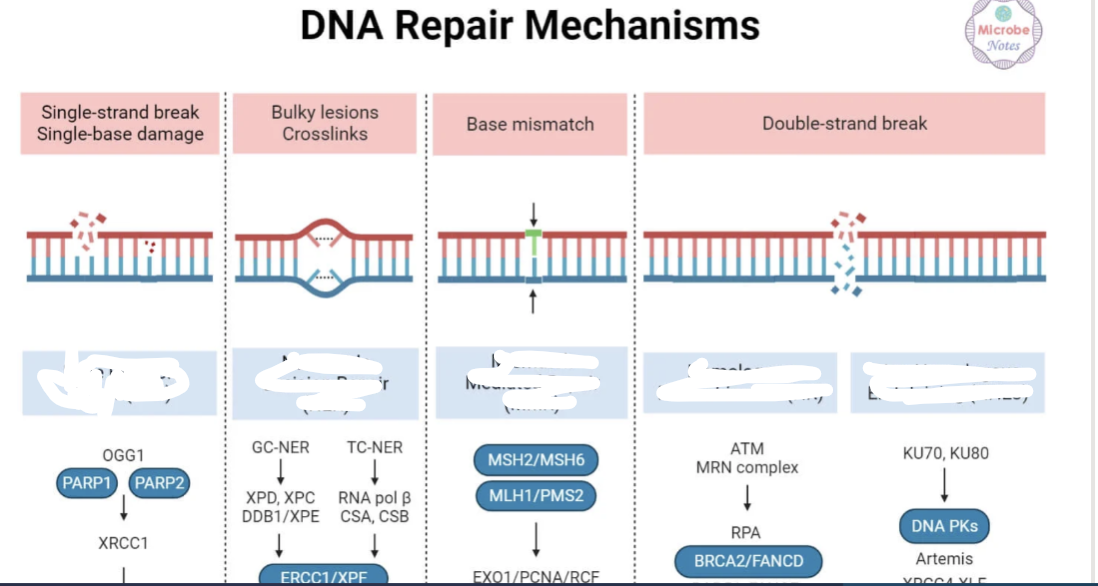
Name the type of repair for the type of damage
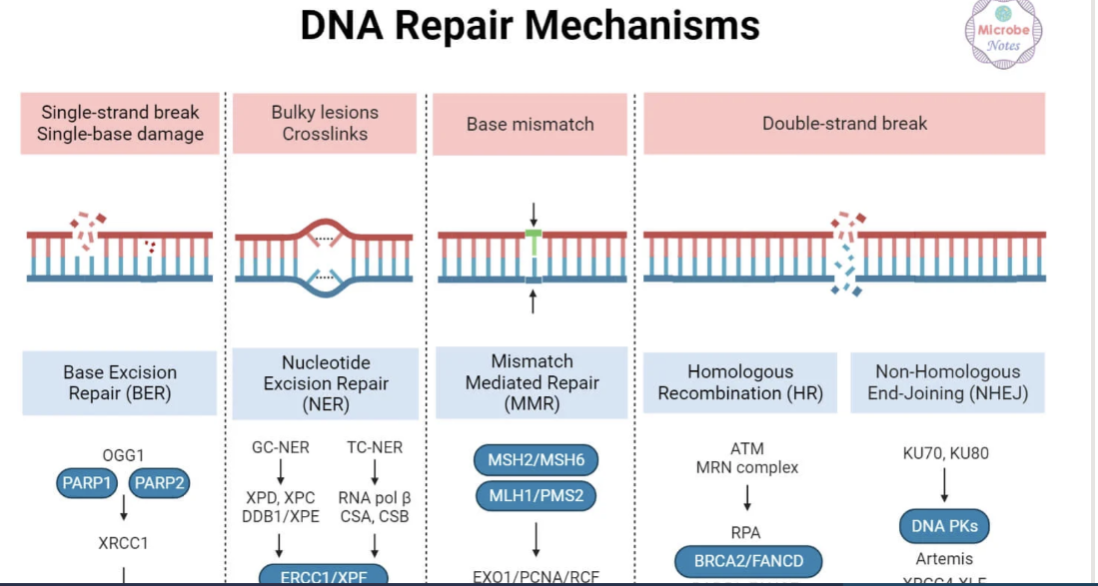
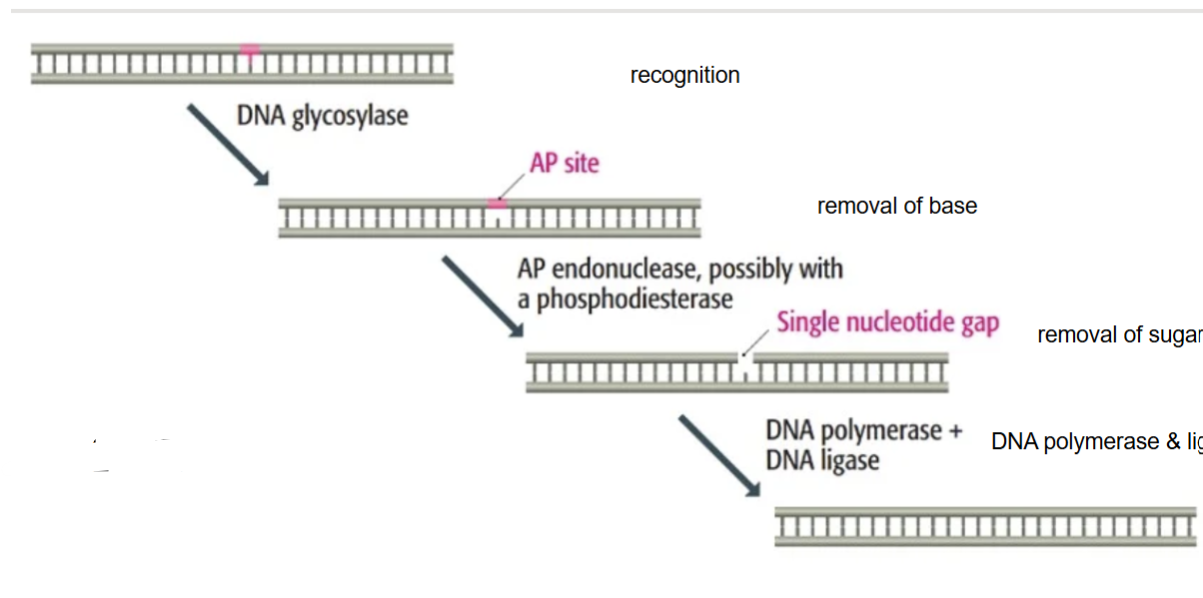
Name this diagram
Base excision repair
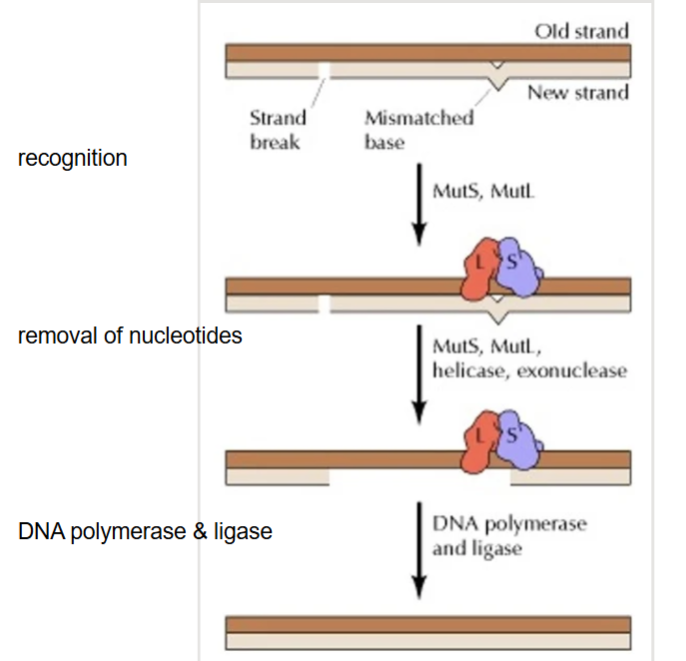
Name this diagram
Mismatch repair
For mismatch repair, how can the cell differentiate the new strand from the template strand?
Cells mark the newly synthesize strand to ensure that they repair the correct strand
• Methylation in prokaryotes
• Nicks in the sugar- phosphate backbone in eukaryotes*
Consequences of mutations: deamination
Unrepaired deamination can lead
to amino acid substitution or a nonsense mutation (creates a truncated protein)
Consequences of mutations: depurination
Unrepaired depurination will
lead to a nucleotide deletion and shift how the remaining information will be read during translation.
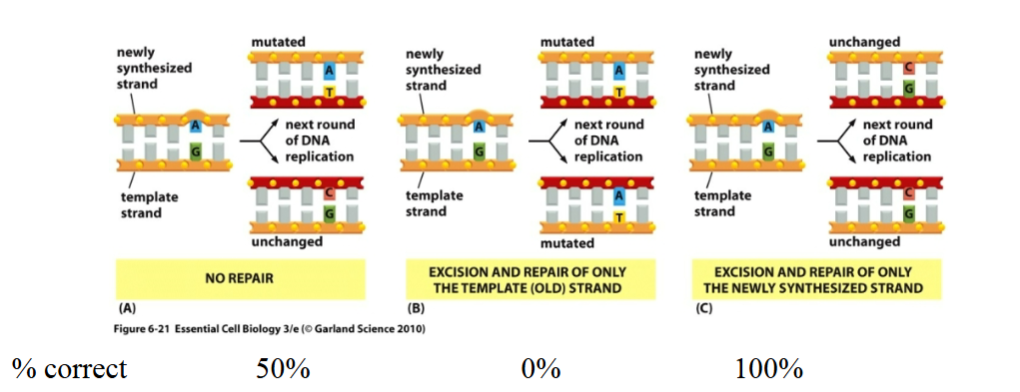
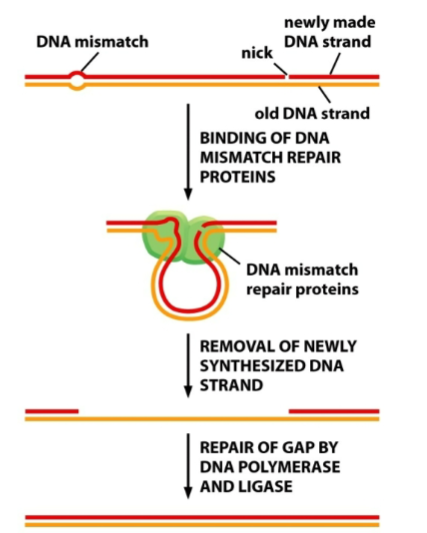
Explain this diagram
DNA mismatch repair...Used for correcting rare mistakes in replication
These outcomes show that cells need the ability to distinguish newly synthesized DNA from the parental template strand (outcome C)
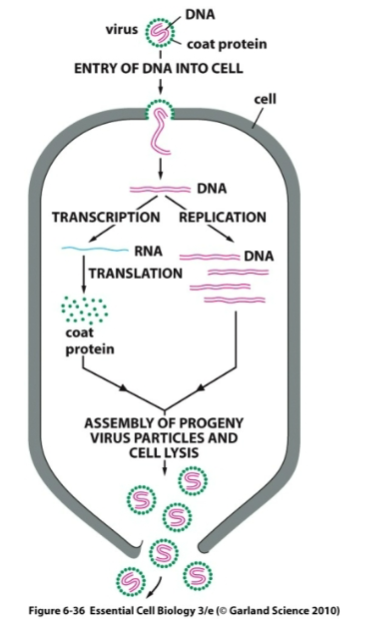
What is this a diagram of
Lytic virus
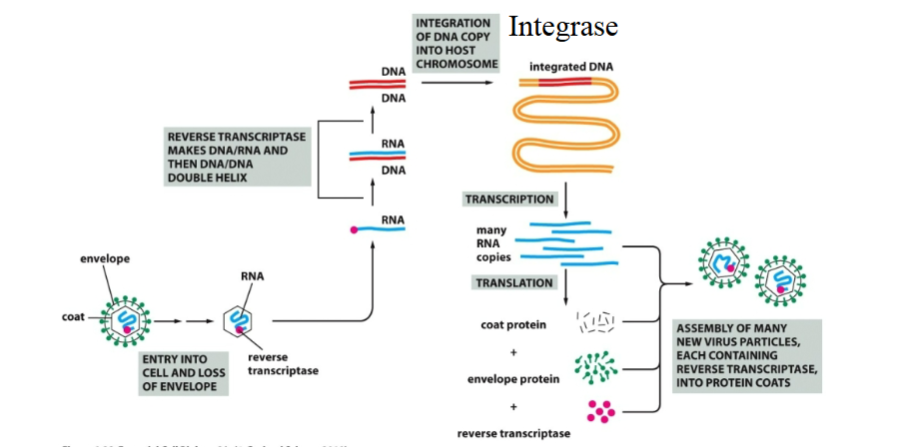
What is this a diagram of
Retroviruses - integration into host as a latent proviruses
two basic types of cells in mammals
• Germ cells - precursor cells that give rise to
gametes. Mutations in germ cells will be passed on
to the offspring.
• Somatic cells - all other cells
Somatic cell mutations will only effect the individual
and not their offspring.
*Mutations in repair genes makes and
individual more susceptible to cancer
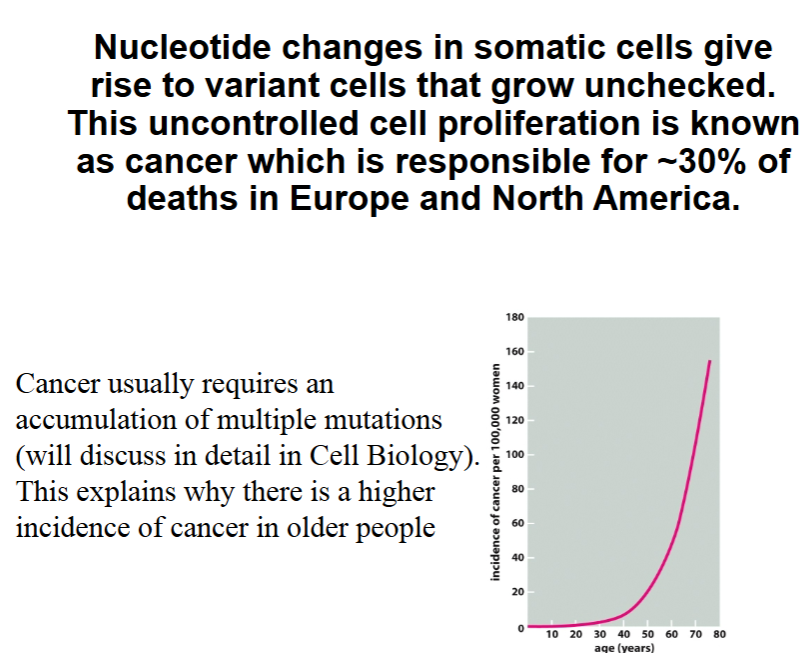
Match the appropriate term to the description:
A consequence of this type of induced mutation is stalling the DNA polymerase
Unrepaired ___________ mutations will lead to a nucleotide deletion in the DNA sequence
You identify a very small region of the liver where your patient’s DNA has a mutation where a cytosine-guanine pair was replaced with an adenosine-thymine pair in 50% of the cells in this small region of the liver. You’ve ruled out mismatch repair through other mechanisms. A _______ mutation is the most likely cause of this type of mutation in the patient’s cells.
DNA alterations that arise within a cell from normal cellular processes, rather than from external factors like radiation or chemicals, are referred to as ___________ DNA damage.
Answer 1
thymidine dimers (thymine dimers)
Answer 2
depurination
Answer 3
deamination
Answer 4
spontaneous
For each of the following statements, select the answer within the brackets that will complete a TRUE statement.
["cut and paste transposons", "retrotransposons"] use an RNA intermediate as part of their transposition process.
Transposition (movement to new location) [does, does not require] sequence similarity to be inserted in a new location within the genome.
Both retroviruses and retrotransposons use a [ ] to create a DNA copy from a mRNA template.
The transposase enzyme from one type of transposon ["can", "can not"] recognize and cut the inverted repeats of another type of transposon.
Answer 1:
retrotransposons
Answer 2:
does not require
Answer 3:
reverse transcriptase
Answer 4:
can not
The sequence below is a region of the top strand of a DNA double helix. If we wanted to use PCR to copy this region of DNA, which of the following primers would be the only set that would work to amplify this DNA region?
top strand: 5′-GCATGCGTGGCATG..........AATTCCATATCCGGAT-3′
["5′-GCATGCGTGG-3' AND 3'-TATAGGCCTA-5'", "3'-CGTACGCACC-5' AND 5'-ATATCCGGAT-3′", "3'-CGTACGCACC-5' AND 5′-GCATGCGTGG-3'"]
The primers for PCR are made of ["RNA nucleotides", "DNA nucleotides"] .
The polymerase will synthesize the copies from ["5' to 3'", "3' to 5'"] .
Answer 1:
5′-GCATGCGTGG-3' AND 3'-TATAGGCCTA-5'
Answer 2:
DNA nucleotides
Answer 3:
5' to 3'
For each of the following statements, select the answer within the brackets that will complete a TRUE statement.
The nucleotide that causes chain termination in dideoxy sequencing has a [ ] at the 3' position of the ribose sugar .
How many primers are need for a sequencing reaction?
The primers used for sequencing are made of [ ] nucleotides
Answer 1:
H at the 3' position of the ribose sugar
Answer 2:
One primer and the polymerase will synthesize the new strand from 5’-3’.
Answer 3:
DNA
A mutation occurs during DNA replication, resulting in a mismatched base pair. The mismatch repair system must identify which strand to correct. Which of the following features would help the system correctly identify the newly synthesized strand? (Select all that apply)
Random selection of the strand to repair
Detection of nicks in the newly synthesized strand
Binding of DNA polymerase to the template strand
Recognition of methylation patterns on the template strand
Detection of nicks in the newly synthesized strand
Recognition of methylation patterns on the template strand
For each of the statements about DNA repair, indicate whether the statement is TRUE or FALSE
["FALSE", "TRUE"] During homologous DNA repair of a double strand break in the paternal chromosome,
the DNA sequences on the maternal chromosome will also be changed.
["FALSE", "TRUE"] Homologous DNA repair requires an RNA primer.
["FALSE", "TRUE"] DNA double strand damage is preferentially repaired using non-homologous DNA repair if the damage occurs after DNA replication but before M-phase.
["TRUE", "FALSE"] The same type of DNA repair polymerase can be used for mismatch repair and for homologous DNA repair.
["FALSE", "TRUE"] Most DNA damage creates structures that are NOT normally seen in DNA.
["TRUE", "FALSE"] Excision Repair is used when only one strand of the helix has DNA damage or nucleotide errors.
Answer 1:
FALSE
Answer 2:
FALSE
Answer 3:
FALSE
Answer 4:
TRUE
Answer 5:
TRUE
Answer 6:
TRUE
For each of the following statements about mutations, select the answer within the brackets that will complete a TRUE statement.
A mutation that results in a single amino acid substitution (different amino acid in the sequence) but has no impact on the protein’s function is a ["silent", "neutral"] mutation.
A DNA mutation that results in a single alteration to the DNA nucleotide and results in the production of a stop codon which terminates protein synthesis prematurely is called a ["silent", "nonsense"] mutation.
Answer 1:
neutral
Answer 2:
nonsense
You read a news story talking about increasing mutation rates observed in the last ten years. A reporter wants to do a follow up on this story, highlighting a family that had a higher than expected incidence of unrepaired thymine dimers but no increases in other types of mutations. The reporter should suspect that this family's accumulation of damage is due to a mutation in __________.
DNA repair polymerase
DNA primase
DNA ligase
the protein that identifies thymine dimers and recruit its nuclease
the protein that identifies thymine dimers and recruit its nuclease