Sequence-Specific Gene control
1/26
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
27 Terms
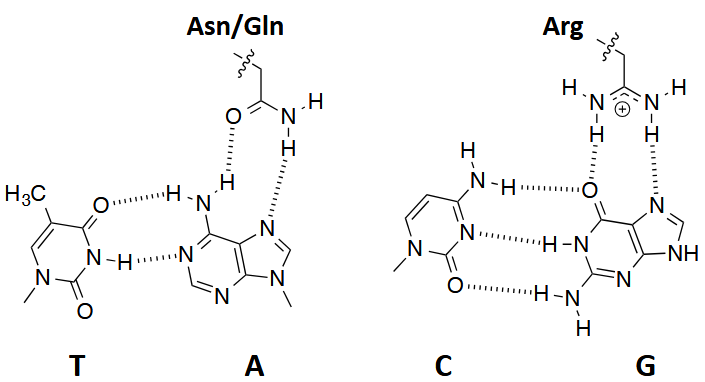
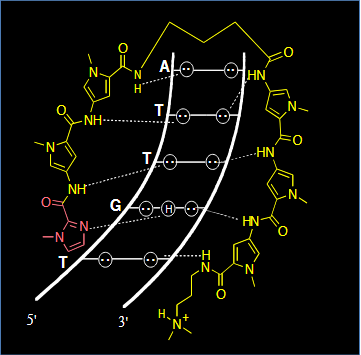
Amino acid – Base interactions
Hoogsteen faces
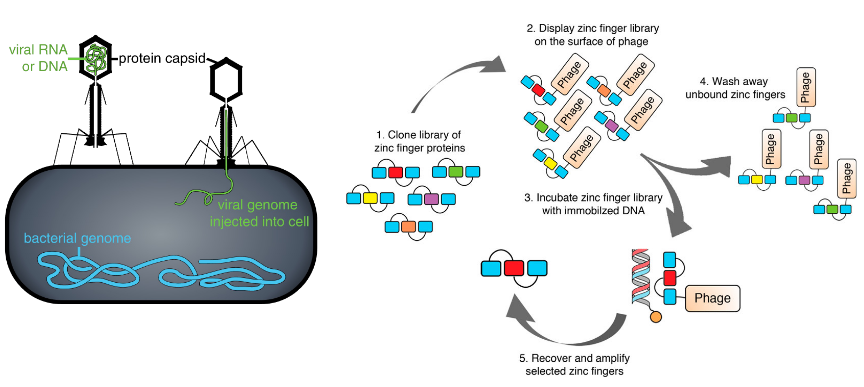
Phage Display
phage: holds DNA inside, expresses proteins outside
DNA-binding experiment
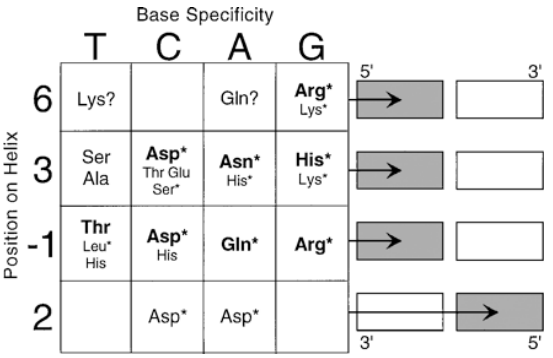
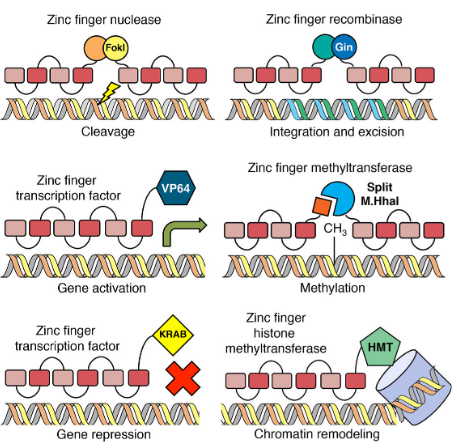
Recognition Code of Zinc fingers
what locations and residues on zinc fingers bind to DNA
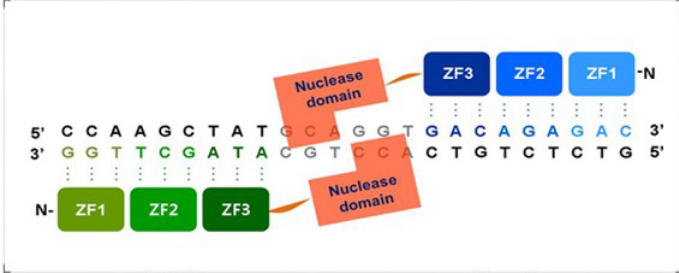
Zinc Finger DNA cleaver
A non-specific nuclease (FokI) borrowed from restriction enzymes is attached to the Zn-Finger DBD; artificial restriction enzymes
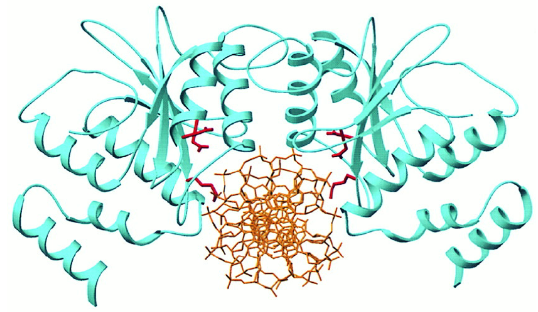
FokI
non-specific nuclease; cleaves phosphodiester groups 9 bp away on the 5′ strand and 13 bp away on the 3′ strand, as indicated by arrows.
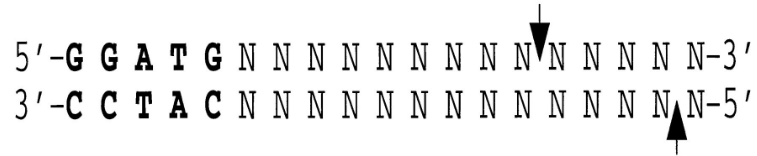
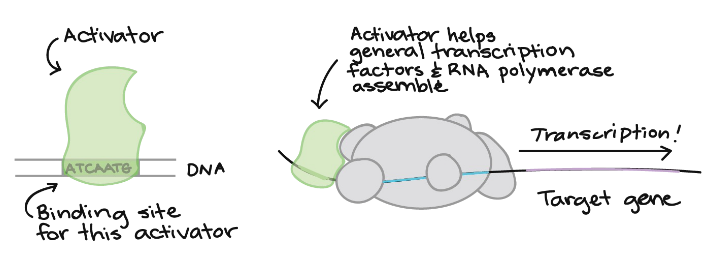
Activation
turns transcription on
Repression
turns transcriptions off
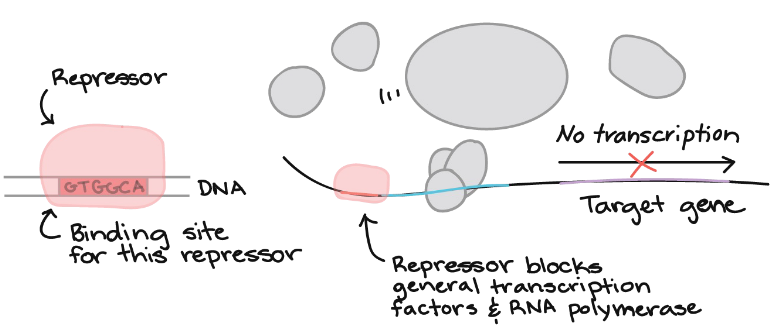
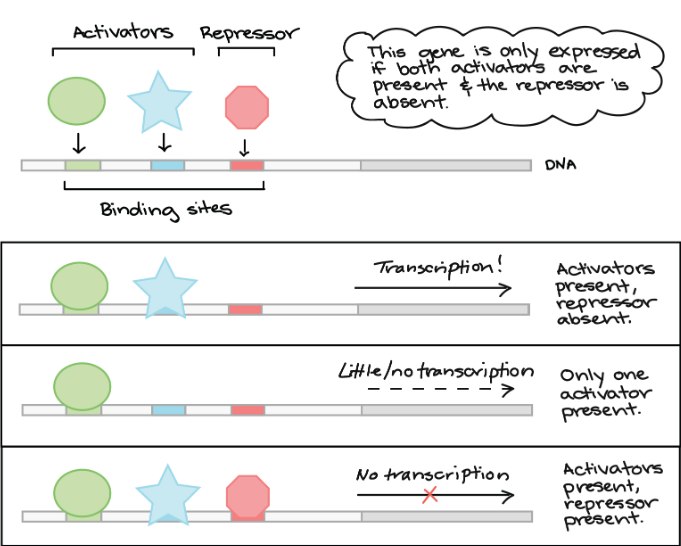
Transcription regulation logic
logic gates
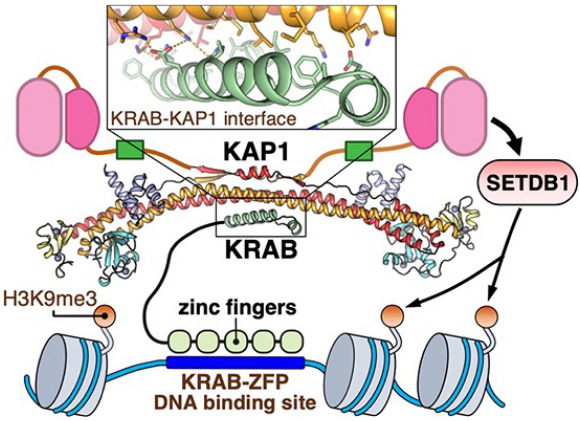
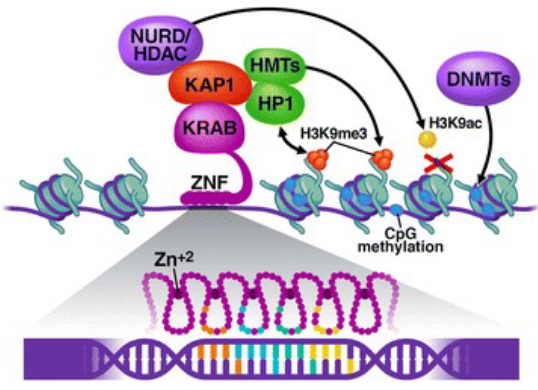
KRAB (Krüppel-associated box)
attached to sequence-specific zinc finger → recruits HMTs → trimethylate histones → silence genes
approximately one third (290) of the 799 different zinc-finger proteins present in the human genome; the largest single family of transcriptional regulators in mammals
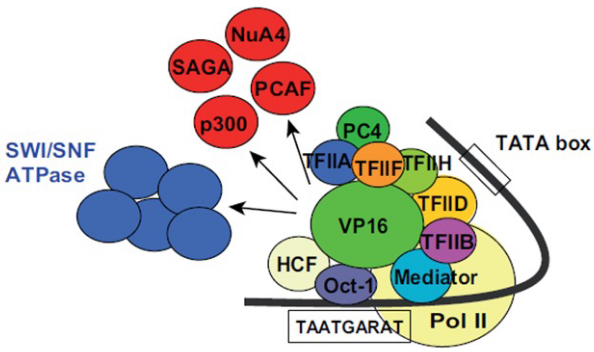
Virus Protein 16 (VP16)
transcription factor from Herpes simplex virus-1 (HSV-1); One of the strongest activation domains known; fused to a DNA-binding domain (DBD) of another protein in order create a sequence-specific artificial transcription activator; facilitates assembly of the preinitiation complex
Zinc finger regulator
add effector domain for site-specfic regulation
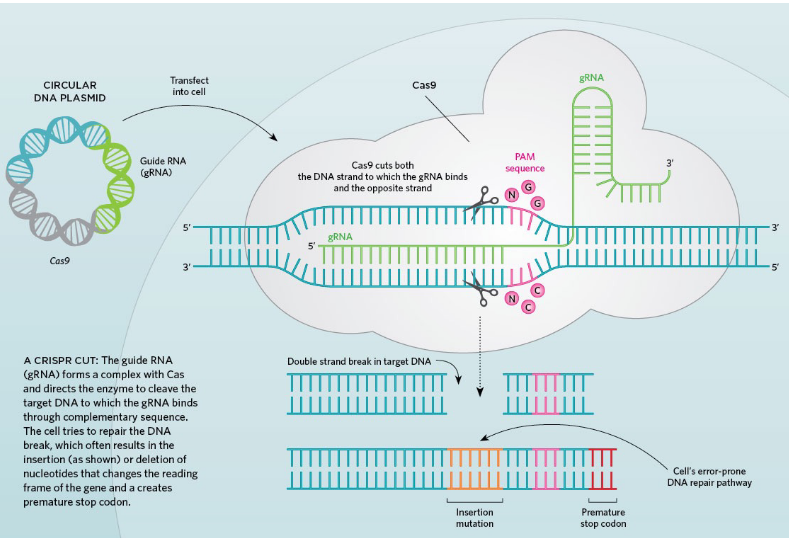
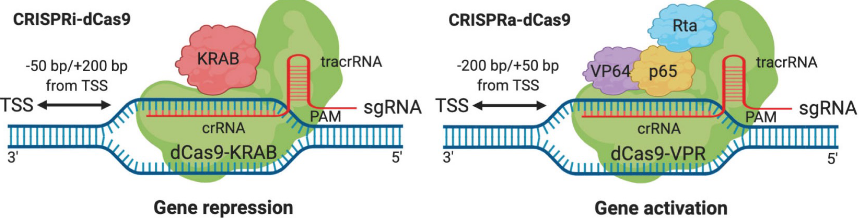
CRISPR/Cas9
plasmid used to alter genome but can be engineered to remove nuclease activity to add regulatory activity
less specific than zinc fingers; but easier to deliver
TALENs (Transcription activator-like effector nuclease)
engineered to bind to practically any desired DNA sequence, so when combined with a nuclease, DNA can be cut at specific locations
sequence-specific targeting issue
Specific targeting of DNA requires recognition of long stretches of DNA; But large proteins and DNA are generally not cell permeable
Small Molecule DNA Binders
intercalators
minor groove binders
major groove binders
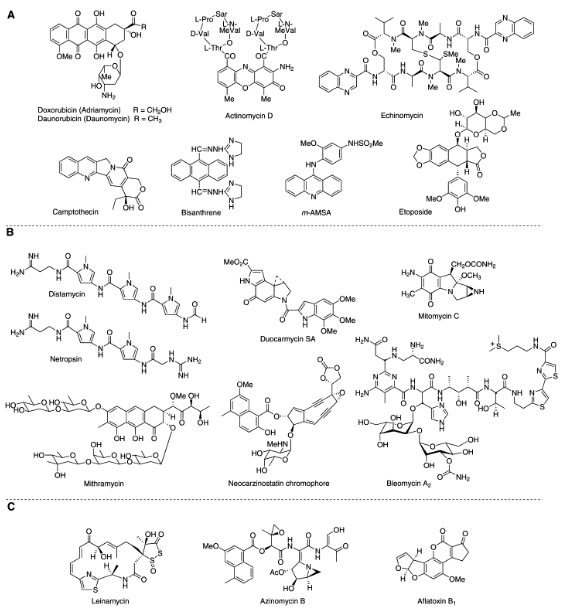
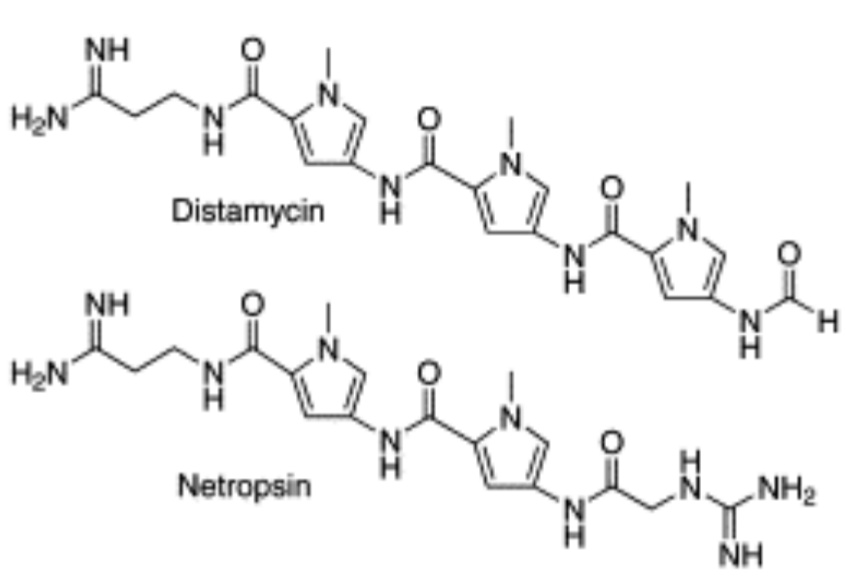
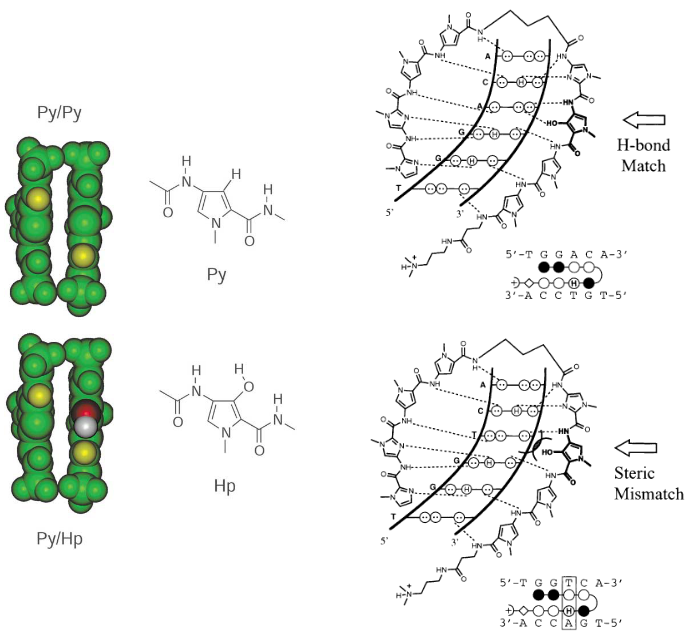
Distamycin/Netropsin
minor groove binders driven by hydrophobic effect
read A-T rich sequences: more flexible, pi-stacking
2:1 dimer stack: possibility to engineer specificity
Rise per Residue of Polyamide Is Similar to Rise of B-DNA
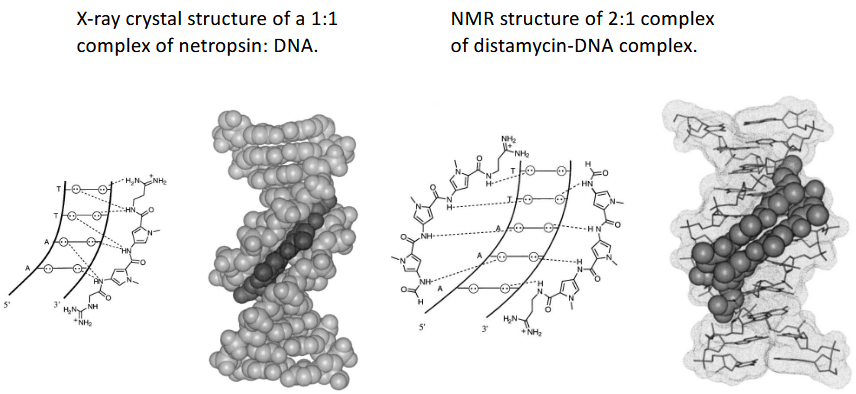
Altered distamycin - C-G/G-C specificity
Imidazole/Pyrrole targets G*C
Pyrrole/Imidazole targets C*G
Pyrrole/Pyrrole targets A*T and T*A
Altered distamycin - lower needed concentration
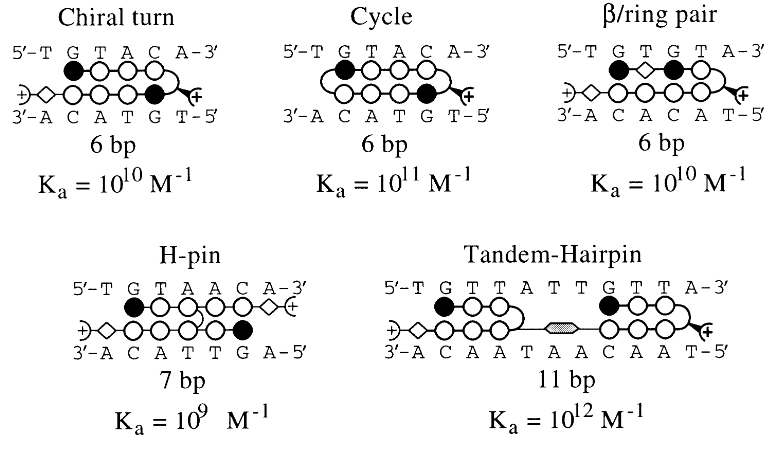
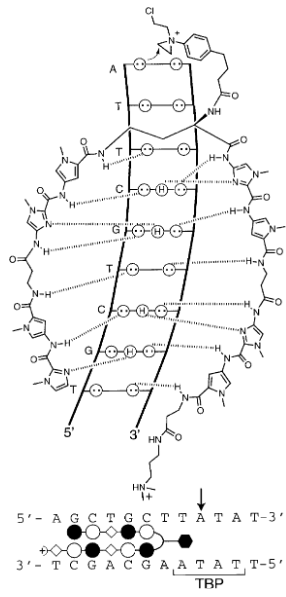
hairpin polyamide motif
Kd = 14 nM
makes molecule larger
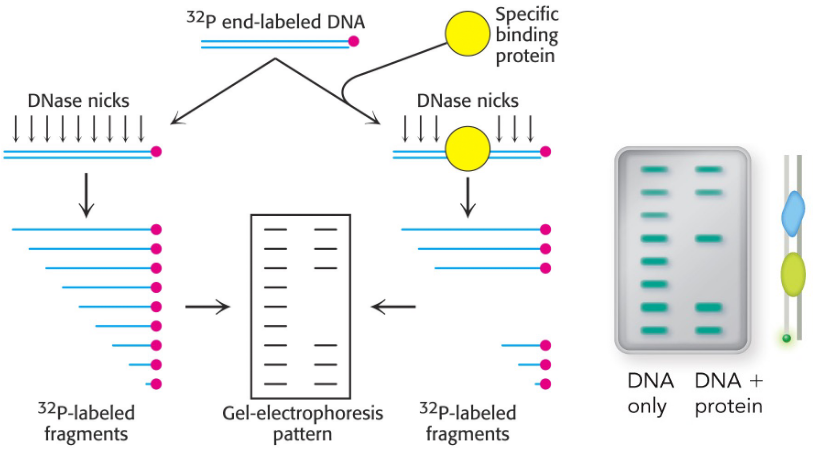
footprinting experiment
where does this molecule bind to DNA?
DNAse randomly cutes DNA but can’t cut where DNA is bound → white space on gel
concentration dependent: 50% concentration = Ka
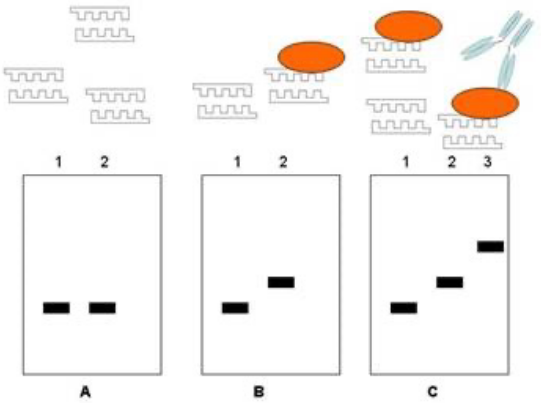
gel shift experiment
NATIVE gel electrophoresis, not denaturing - double strand
finds binding affinity - concentration dependent
molecule binding motifs and affinity
Altered distamycin - A-T vs T-A discrimination
use hydroxy pyrole
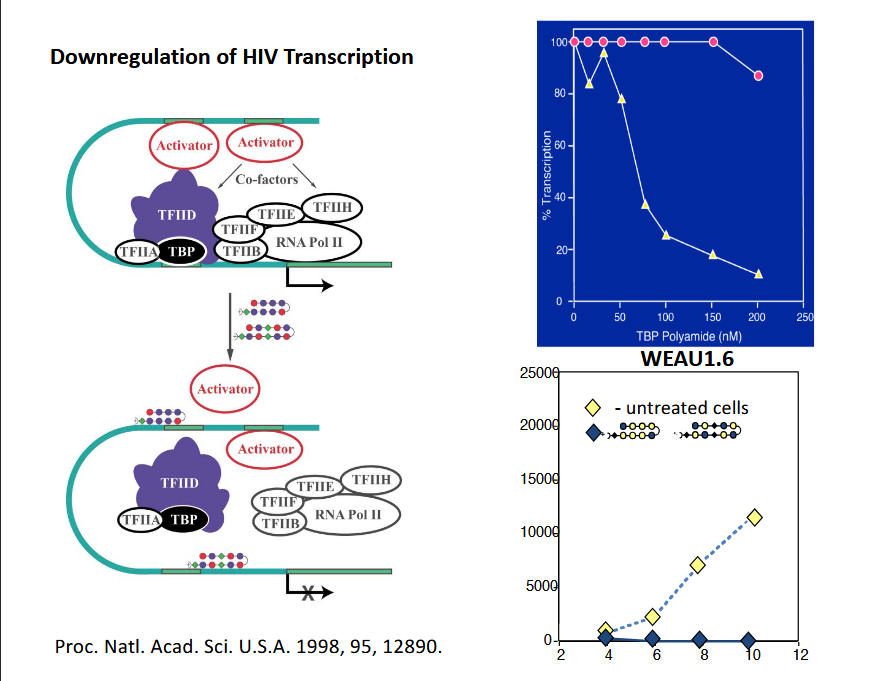
Downregulation of HIV Transcription
hairpin binders prevent binding to transciptional zones
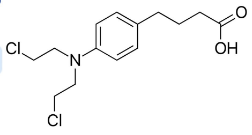
Chlorambucil
mustard gas reactivity
animal chemotherapeutic
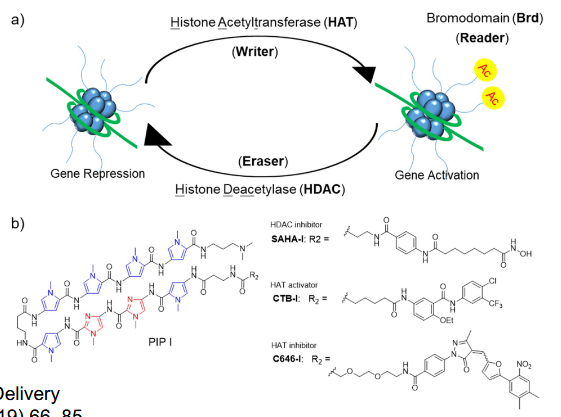
Regulation by HDAC Inhibitor
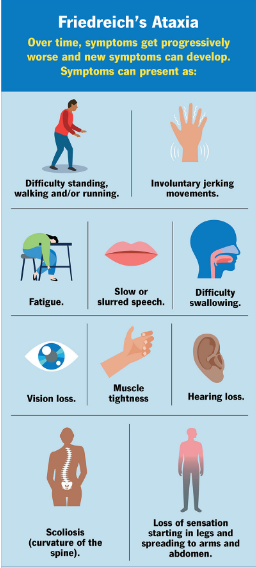
Friedreich’s Ataxia
an inherited neurodegenerative disorder that impacts children
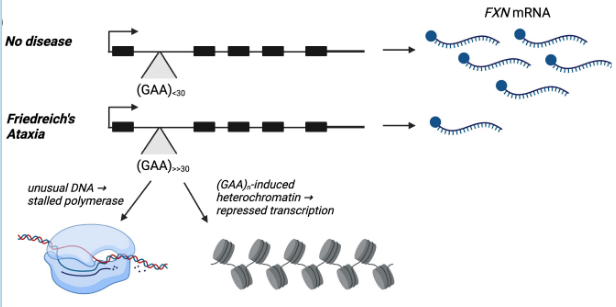
triple-repeat neurological disorder → repeats cause folding, more heterochromatin, and stalled transcription → less mRNA made
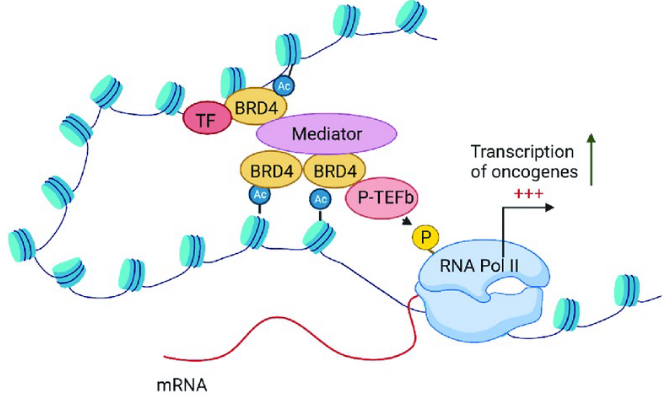
BRD4
passive scaffold protein and active kinase that phosphorylates RNA polymerase II
possible therapeutic for Friedreich’s Ataxia → turn on transcription