Cell Bio Exam 2 Prep
1/114
Earn XP
Description and Tags
(ch. 16-19)
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
115 Terms
The information encoded in DNA is used within a cell in a two-stage process. The two stages
of this process are called
A) denaturation and renaturation.
B) base-pairing and denaturation.
C) replication and division.
D) transcription and translation.
E) replication and transcription.
D) transcription and translation.
2) Which of the following model organisms were used in experiments demonstrating that DNA
was the genetic material?
A) mice
B) amoeba
C) humans
D) tobacco plants
E) bacteria and bacteriophages
E) bacteria and bacteriophages
3) The scientist credited with the discovery of DNA (nuclein) from white blood cells in pus on
surgical bandages is
A) Johann Friedrich Meischer.
B) Eduard Zacharias.
C) Fredrick Griffiths.
D) Martha Chase.
E) Oswald Avery.
A) Johann Friedrich Meischer.
4) In 1928, Frederick Griffith's experiments with Streptococcus pneumoniae suggested that
A) bacteria do not incorporate the DNA of other bacteria.
B) attenuated R (rough) bacteria easily revert to the wild type (virulent).
C) RNA was the genetic material.
D) heat-killed bacteria could somehow "transform" live bacteria.
E) protein was the genetic material.
D) heat-killed bacteria could somehow "transform" live bacteria.
5) Following reports that the amount of DNA varied over time within cells, scientists in the early
1900s concluded that the genetic material had to be
C) protein.
6) In an experiment like that of the 1952 Hershey and Chase experiments, 35S was added to a
phage replicating within its bacterial host. The new phage particles were carefully isolated and
used to infect fresh bacterial cells in the absence of any radioisotopes. Where would you expect
to find the 35S radioisotope immediately after infection?
A) in the phage ghosts outside the bacterial cells
B) incorporated into the bacterial matrix proteins
C) incorporated into the bacterial DNA
D) inside the bacterial cells, but separate from the bacterial DNA
E) inside the bacterial cells, but separate from the bacterial proteins
A) in the phage ghosts outside the bacterial cells
) Avery, MacLeod, and McCarty (1944) demonstrated that DNA was the genetic material by
treating heat-killed Streptococcus pneumoniae S (smooth) strain with DNase, thereby preventing
transfer of the "transforming substance" from the killed S strain to the live R (rough) strain when
the two were mixed together and injected into mice. Another way that one could demonstrate
transformation in bacteria would be to extract DNA from
A) an R strain and mix it with cells of an S strain.
B) both S and R strains and mix it to allow recombination to take place.
C) an ampicillin-sensitive strain and mix it with cells of an ampicillin-resistant strain.
D) an S strain and mix it with cells of an R strain.
E) None of the above are correct.
D) an S strain and mix it with cells of an R strain.
8) Which of the following are components of Chargaff's rules of bases?
A) % purines = % pyrimidines
B) %C + %T = %A + %G
C) %A = %T
D) %G = %C
E) All of these are true.
E) All of these are true.
9) An example of a purine is
A) thymine.
B) guanine.
C) deoxyribose.
D) cytosine.
E) ribose 5-phosphate.
B) guanine.
10) When examining a five-carbon sugar to determine if it is ribose or deoxyribose, one looks to
see if there are two H on the ________ carbon in the structure.
A) 1'
B) 2'
C) 3'
D) 4'
E) 5'
B) 2'
11) DNA is different from RNA in that
A) RNA contains an additional oxygen atom on the ribose sugar.
B) RNA is made up five bases, whereas DNA is made up of four.
C) RNA cannot exist as a double helix.
D) in general, RNA molecules are longer than DNA molecules.
E) All of these statements are true.
A) RNA contains an additional oxygen atom on the ribose sugar.
12) The 5' end of a DNA molecule can be chemically distinguished from the 3' end because there
is a(n) ________ group at the 5' end and a(n) ________ group at the 3' end.
A) amine; carboxyl
B) phosphate; sulfate
C) hydroxyl; hydrogen
D) phosphate; hydroxyl
E) amine; hydroxyl
D) phosphate; hydroxyl
13) The base composition in DNA isolated from cow liver cells is 28% adenine; what percent of
the bases are cytosine?
A) 14%
B) 22%
C) 28%
D) 36%
E) 56%
B) 22%
14) DNA isolated from Aspergillis has an adenine content of 25%. Based upon this information,
what is the %G + %C within the Aspergillis DNA?
A) 0%
B) 25%
C) 50%
D) 75%
E) The answer cannot be determined from this information.
C) 50%
15) Which of the following shows Chargaff's equivalence?
A) RNA
B) single-stranded DNA
C) histones
D) double-stranded DNA
E) deoxyribose
D) double-stranded DNA
16) Which of the following statements regarding the Watson-Crick model of DNA is false?
A) It is the most common biological form of DNA.
B) The two DNA strands form a left-handed helix.
C) The two DNA strands are antiparallel.
D) The bases are "stacked" in the center of the molecule.
E) The backbone consists of alternating sugars and phosphates.
B) The two DNA strands form a left-handed helix.
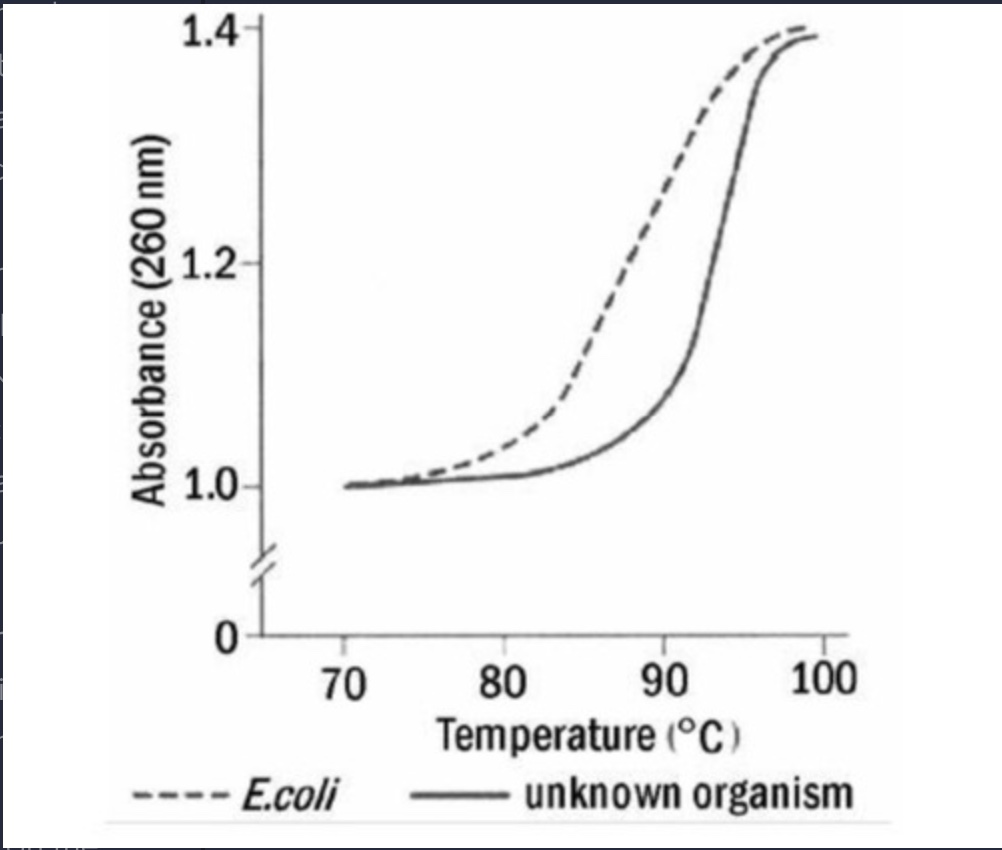
17) A recently isolated bacterial strain has been given to you for general characterization of the
genome. You begin by isolating the DNA and subjecting it to thermal denaturation. You use
Escherichia coli (E. coli) DNA as a reference.
Based on the information in the graph, you can determine that the DNA of the new strain has a
high
A) G-C content with respect to E. coli.
B) pyrimidine content with respect to E. coli.
C) A-T content with respect to E. coli.
D) purine content, with respect to E. coli.
E) histone content with respect to E. coli.
A) G-C content with respect to E.
18) Circular DNA molecules found in nature are
A) positively supercoiled.
B) negatively supercoiled.
C) not supercoiled, due to their small size.
D) always oriented in a triple helix to conserve space.
E) always in the Z-DNA formation.
B) negatively supercoiled.
19) What characteristic is shared between most prokaryotic and mitochondrial genomes?
A) <5 % protein-coding genes
B) >20% interspersed repeats
C) circular chromosomes
D) linear chromosomes
E) circular chromosomes with <5 % protein-coding genes
C) circular chromosomes
20) Mitochondrial and chloroplast genomes typically
A) are composed of primarily noncoding DNA.
B) encode all of the proteins needed for aerobic respiration and photosynthesis, respectively.
C) encode for ribosomal and transfer RNAs.
D) encode some of the proteins needed for aerobic respiration and photosynthesis, respectively.
E) encode some of the proteins needed for aerobic respiration and photosynthesis, respectively,
as well as ribosomal and transfer RNA.
E) encode some of the proteins needed for aerobic respiration and photosynthesis, respectively,
as well as ribosomal and transfer RNA.
21) Heterochromatin is highly ________, thus constitutive heterochromatin plays a(n) ________role and facultative heterochromatin functions in ________.
A) compacted; structural; regulation of gene expression
B) compacted; regulatory; structure
C) denatured; regulatory; structure
D) denatured; structural; regulation of gene expression
E) modified; evolutionary; structure
A) compacted; structural; regulation of gene expression
22) Alteration of histones by addition of methyl and acetyl groups to particular amino acids
A) changes B-DNA to Z-DNA.
B) results in the remodeling of chromatin that can activate or inhibit gene expression.
C) creates G bands seen after Giemsa staining of chromosomes.
D) helps preserve telomeres during DNA replication.
E) alters the denaturation and renaturation of DNA.
B) results in the remodeling of chromatin that can activate or inhibit gene expression.
23) Fluorescent in situ hybridization (FISH) would be helpful in determining
A) the location of B-DNA and Z-DNA.
B) the position and structure of nucleosomes.
C) the position of particular chromosomes or sequences inside the nucleus.
D) the base composition of a DNA sample.
E) the number of nuclear pores on the nuclear envelope
C) the position of particular chromosomes or sequences inside the nucleus.
24) Long interspersed nuclear elements (LINEs) found in eukaryotic genomes
A) are mobile pieces of DNA called transposons that can copy themselves and move around the
genome.
B) are thought to create genomic variability important to evolutionary adaptation.
C) are repeated 10-1000 times in clusters near centromeres and telomeres.
D) are part of constitutive heterochromatin.
E) are mobile pieces of DNA called transposons that can copy themselves and move around the
genome, and they are thought to create genomic variability important to evolutionary adaptation.
E) are mobile pieces of DNA called transposons that can copy themselves and move around the
genome, and they are thought to create genomic variability important to evolutionary adaptation.
25) Which type of DNA makes up the largest portion of the human genome?
A) introns
B) unique noncoding DNA
C) exons
D) tandemly repeated DNA
E) interspersed repeated DNA
E) interspersed repeated DNA
26) Interspersed repeated DNA
A) constitutes 25%-40% of most mammalian genomes.
B) is located in one region of most mammalian genomes.
C) is approximately 105-107 bp in length for each repeat.
D) can be classified into gene families, such as the Alu family.
E) constitutes 20%-40% of most mammalian genomes and can be classified into gene families,
such as the Alu family.
E) constitutes 20%-40% of most mammalian genomes and can be classified into gene families,
such as the Alu family.
27) Which type of DNA makes up the smallest portion of the human genome?
A) introns
B) unique noncoding DNA
C) exons
D) tandemly repeated DNA
E) interspersed repeated DNA
C) exons
28) Packaging of prokaryotic chromosomes
A) doesn't involve histone or histone-like proteins.
B) involves large regions of positively supercoiled DNA but lacks histones or histone-like
proteins.
C) is very simple and lacks higher-level structure, such as looped domains.
D) involves structural RNA molecules that contribute to the formation of looped domains. is
essentially the same as eukaryotic DNA packaging.
D) involves structural RNA molecules that contribute to the formation of looped domains. is
essentially the same as eukaryotic DNA packaging.
29) Which of the following statements is true regarding mitochondrial and chloroplast genomes?
A) They contain mostly interspersed and tandemly repeated DNA.
B) They are able to replicate independently of the nuclear DNA.
C) They contain all the genes necessary for aerobic respiration.
D) They are degenerate bacterial genomes that don't contain important protein-coding genes.
E) They only encode for ribosomal and transfer RNAs.
B) They are able to replicate independently of the nuclear DNA.
30) Which of the following is the correct order of the levels of DNA packaging in eukaryotic
chromosomes?
A) nucleosome → chromatin fiber → looped domains → heterochromatin
B) nucleosome → looped domains → chromatin fiber → heterochromatin
C) heterochromatin → nucleosome → chromatin fiber → looped domains
D) chromatin fiber → heterochromatin → nucleosome → looped domains
E) chromatin fiber → nucleosome → looped domains → heterochromatin
A) nucleosome → chromatin fiber → looped domains → heterochromatin
31) Unusual eukaryotic-like (possessing a defined nucleus) cells were obtained from a sample of
water from a subglacial antarctic lake. To investigate the properties of the new organism's
genome, the nucleus was carefully isolated and the chromatin obtained. The fibers had a "beads
on a string" appearance, suggesting some type of "histones." Following digestion with
micrococcal nuclease, protein removal, and gel electrophoresis, DNA fragments were obtained
that are multiples of 280 base pairs in length. Longer incubation with micrococcal nuclease prior
to protein removal results in fragments 200 base pairs in length. The amount of DNA that is
wrapped around the "core histones" is therefore
A) 80 base pairs in length.
B) 200 base pairs in length.
C) 360 base pairs in length.
D) 480 base pairs in length.
E) Not enough information is given to determine the answer.
B) 200 base pairs in length.
32) The nuclear envelope functions as a
A) means of separating nuclear and cytoplasmic constituents.
B) means of localization of the chromosomes within the cell.
C) selective barrier, allowing certain substances in or out.
D) means of sequestering many of the mRNA processing activities from the cytosol.
E) selective barrier that separates nuclear and cytoplasmic constituents, localizes chromosomes
within the cell, and sequesters many of the mRNA processing activities from the cytosol.
E) selective barrier that separates nuclear and cytoplasmic constituents, localizes chromosomes
within the cell, and sequesters many of the mRNA processing activities from the cytosol.
33) You are using genetic engineering to design a protein that needs to be synthesized in the
cytoplasm and then localized in the nucleus of the cell. To accomplish this, you could add
A) a DNA sequence for a nuclear localization signal into the DNA that will be present in the
mature protein.
B) the protein during mitosis, at a time when the nuclear membrane is dispersed.
C) a nucleotide or few nucleotides to the DNA sequence for the protein.
D) a series of specific polysaccharides to the carboxyterminal end of the protein.
E) inverted repeats to the DNA sequence for the protein.
A) a DNA sequence for a nuclear localization signal into the DNA that will be present in the
mature protein.
34) Numerous Xenopus cells were exposed to varying amounts of ultraviolet light. Some of the
resulting cells were observed to lack nucleoli. As a result, one would expect that the cell could
not
A) perform cellular respiration.
B) replicate its DNA.
C) perform photosynthesis.
D) synthesize ribosomal RNA.
E) perform cellular respiration or replicate its DNA.
D) synthesize ribosomal RNA.
35) The nucleolus
A) is surrounded by a thin nucleolar membrane.
B) is found in prokaryotes.
C) contains tRNA.
D) synthesizes ribosomal RNA.
E) is found in prokaryotes and contains tRNA and rRNA.
D) synthesizes ribosomal RNA. key said C but internet says otherwise
36) Which of the following structures contains the receptor protein importin?
A) mitochondrion
B) nucleolus
C) chloroplast
D) plastid
E) nucleus
E) nucleus
37) Mitochondrial DNA can be used to identify a species based upon
A) NADH dehydrogenase sequences.
B) variation in the mitochondrial rRNA genes.
C) the 648 bp DNA bar code region.
D) the proportion of GC base pairs.
E) the size of the mitochondrial genome.
C) the 648 bp DNA bar code region.
38) Each of the following structures is found within the nucleus of a eukaryotic cell except
A) transfer RNA.
B) histones.
C) plasmids.
D) the nucleolus.
E) All of these are found in the nucleus.
C) plasmids.
1) The DNA polymerase reaction occurs in the ________ as incoming nucleotides are added to
________ end(s) of the growing DNA chain.
A) 5' → 3' direction; the 3'-hydroxyl
B) 5' → 3' direction; the 3'-phosphate
C) 3' → 5' direction; the 5'-hydroxyl
D) 3' → 5' direction; the 5'-phosphate
E) 5' → 3' direction or 3' → 5' direction; both
A) 5' → 3' direction; the 3'-hydroxyl
2) The energy needed to polymerize DNA is supplied by
A) the hydrolysis of the phosphate bonds of ATP by an accessory protein that is part of the DNA
polymerase holoenzyme.
B) the hydrolysis of the phosphate bonds of ATP or GTP, depending on the form of DNA
polymerase.
C) the hydrolysis of the phosphate bonds of the deoxynucleoside triphosphates that are being
added to the new DNA chain.
D) the formation of the covalent bond between the phosphate of the nucleoside and the hydroxyl
group.
E) the hydrolysis of the RNA primers used in DNA replication.
C) the hydrolysis of the phosphate bonds of the deoxynucleoside triphosphates that are being
added to the new DNA chain.
3) In DNA replication, the leading strand of DNA ________, while the other newly forming
strand, called the lagging strand, ________.
A) is synthesized in the 5' → 3' direction; the new DNA is synthesized in the 3' → 5' direction
B) is synthesized in the 3' → 5' direction; the new DNA is synthesized in the 5' → 3' direction
C) does not require an RNA primer; an RNA primer is required
D) is synthesized as a continuous chain; the new DNA is formed in a series of short,
discontinuous fragments
E) is synthesized in a series of short, discontinuous fragments; the new DNA is synthesized as a
continuous chain
D) is synthesized as a continuous chain; the new DNA is formed in a series of short, discontinuous fragments
4) The most common form of spontaneous replication error is
A) tautomeric shift.
B) double-stranded DNA breaks.
C) thymine dimer addition.
D) base analogue addition.
E) DNA intercalation.
A) tautomeric shift.
5) Strand slippage results in ________, which can cause a number of human diseases such as
fragile X syndrome.
A) tautomeric shift
B) double-stranded DNA breaks
C) pyrimidine dimer addition
D) deletion of large sections of DNA
E) the re-replication of highly repetitive DNA sequences
E) the re-replication of highly repetitive DNA sequences
6) DNA replication is
A) conservative.
B) dispersive.
C) semiconservative.
D) irregular.
E) None of these selections apply.
C) semiconservative.
7) Which of the following do not undergo theta replication?
A) bacterial genomes
B) mitochondrial genomes
C) chloroplast genomes
D) plasmids
E) protist genomes
E) protist genomes
8) DNA replication
A) requires a type of RNA polymerase.
B) is partially regulated by promoter/terminator sites.
C) proceeds by making two continuous strands.
D) is not edited once polymerization has occurred.
E) is conservative.
A) requires a type of RNA polymerase
9) Each of the following are associated with replicons except
A) origin of replication.
B) terminator sites.
C) initiator proteins.
D) minichromosome maintenance proteins.
E) helicase loaders.
B) terminator sites.
10) A NASA probe returned samples of rocks from the surface of Mars. From these rocks, a
prokaryotic organism was isolated and remained viable under conditions similar to those of
Mars. The organisms are capable of dividing to replicate themselves. In order to determine
whether the DNA replication of these organisms was semiconservative, an experiment using
N15/N14 (similar to that of Meselson and Stahl) was performed. If the method of DNA
replication were conservative, after one generation one would expect to see
A) all of the DNA in the heavy (N15) band.
B) all of the DNA in the light (N14) band.
C) all of the DNA at a point midway between the heavy and light bands.
D) half of the DNA in the light band, the other half in the heavy band.
E) None of these are correct.
D) half of the DNA in the light band, the other half in the heavy band.
11) An isolate of a mutant bacterium appears to grow more slowly than the wild-type population
from which it was isolated. Further studies showed that the slower growth was due to a markedly
reduced DNA polymerase I activity. From this information, one would expect that this organism
would also be deficient in the activity of DNA
A) excision repair.
B) recombination.
C) transcription.
D) translation.
E) unwinding.
A) excision repair.
12) Spontaneous hydrolysis reactions between DNA and surrounding water molecules
commonly cause
A) pyrimidine dimer formation.
B) depurination.
C) tautomeric shift.
D) double-stranded DNA breaks.
E) deletion mutations.
B) depurination.
13) Deamination is a spontaneous DNA mutation that converts cytosine to
A) thymine.
B) guanine.
C) adenine.
D) uracil.
E) None of these are correct.
d. uracil
14) Thymine is used in DNA despite the fact that it is energetically more expensive to synthesize
than uracil because
A) spontaneous deamination reactions convert cytosine to uracil at a fairly high rate.
B) uracil is more sensitive to ultraviolet (UV) damage.
C) uracil is more susceptible to depurination.
D) uracil can only bind with ribose, not deoxyribose.
E) thymine is less likely to undergo spontaneous deamination than uracil
A) spontaneous deamination reactions convert cytosine to uracil at a fairly high rate
15) Ultraviolet (UV) light-treated bacteria would have increased DNA damage in the form of
A) analog incorporation.
B) pyrimidine dimer formation.
C) intercalation of the bases.
D) direct transition of the bases.
E) deamination.
B) pyrimidine dimer formation.
16) DNA damage can occur
A) due to exposure to certain chemicals or radiation.
B) spontaneously.
C) during DNA replication.
D) during transcription.
E) during DNA replication, spontaneously, or due to exposure to certain chemicals or radiation.
E) during DNA replication, spontaneously, or due to exposure to certain chemicals or radiation.
17) Base-modifying chemicals such as aflatoxin B1 or nitrosoguanidine cause mutations by
A) increasing the likelihood of depurination.
B) causing double-stranded breaks in the DNA sugar-phosphate backbone.
C) causing mispairing during DNA replication.
D) causing pyrimidine dimers.
E) increasing the likelihood of depurination or causing mispairing during DNA replication.
E) increasing the likelihood of depurination or causing mispairing during DNA replication.
18) Insertion and deletion mutations can be caused by
A) spontaneous hydrolysis reactions between DNA and water.
B) intercalating agents.
C) the addition of DNA adducts.
D) tautomeric shift.
E) spontaneous hydrolysis reactions between DNA and water or the addition of DNA adducts.
B) intercalating agents.
19) DNA repair in eukaryotes and prokaryotes
A) relies on DNA polymerases to correct all mutations.
B) relies on completely different mechanisms.
C) is accomplished by multiple specialized pathways.
D) can only repair abnormal nucleotides such as pyridine dimers and depurinated bases.
E) is very ineffective.
C) is accomplished by multiple specialized pathways
20) Which of the following proteins is observed exclusively in association with prokaryotic
DNA replication?
A) DNA polymerase I
B) primase
C) helicase
D) single-strand binding proteins
E) telomerase
A) DNA polymerase I
21) Which of the following eukaryotic DNA polymerases replicates mitochondrial DNA?
A) alpha
B) beta
C) delta
D) epsilon
E) gamma
E) gamma
22) Which of the following proteins is observed exclusively in association with eukaryotic DNA
replication?
A) DNA polymerase I
B) DNA gyrase
C) single-strand binding proteins
D) DNA ligase
E) telomerase
E) telomerase
23) Base excision repair is used to
A) correct base mismatches in newly synthesize DNA.
B) fix double-strand DNA breaks.
C) remove intercalating agents.
D) remove modified or depurinated bases.
E) remove intercalating agents and modified or depurinated bases.
D) remove modified or depurinated bases.
24) Nonhomologous end joining is used to
A) correct base mismatches in newly synthesized DNA.
B) fix double-strand DNA breaks.
C) avoid deletion or insertion of bases.
D) remove modified or depurinated bases.
E) avoid deletion or insertion of bases and remove modified or depurinated bases.
B) fix double-strand DNA breaks.
25) A 40-year-old patient has been diagnosed with cancer. Sequencing of cancer cell genomes
reveals a high frequency of G/C to A/T base pairs conversion. The DNA repair system that is
most likely deficient in this patient is the
A) base excision repair pathway.
B) SOS repair pathway.
C) nonhomologous end-joining pathway.
D) nucleotide excision repair pathway.
E) synthesis-dependent strand annealing pathway.
A) base excision repair pathway.
26) Cells that are very sensitive to UV radiation most likely have a defect in the
A) base excision repair pathway.
B) SOS repair pathway.
C) nonhomologous end-joining pathway.
D) nucleotide excision repair pathway.
E) synthesis-dependent strand annealing pathway.
D) nucleotide excision repair pathway.
27) In hereditary nonpolyposis colon cancer, mutations in MutS and MutH proteins result in the
accumulation of mutations caused by
A) mismatched base pairs.
B) structurally damaged bases.
C) double-stranded DNA breaks.
D) thymine dimers.
E) DNA recombination.
A) mismatched base pairs.
28) Mismatch repair targets improperly paired nucleotides. In prokaryotes, the incorrect member
of an abnormal base pair can be differentiated from the correct member because
A) the incorrect member has undergone a hydroxylation reaction.
B) newly synthesized DNA contains uracil.
C) the incorrect member doesn't match the surrounding bases.
D) the original strand of DNA contains methylated bases while newly synthesized DNA does
not; thus the correct member is in the methylated strand.
E) the correct member has been deaminated.
D) the original strand of DNA contains methylated bases while newly synthesized DNA does not; thus the correct member is in the methylated strand.
29) SOS repair and translesion synthesis employ
A) repair endonucleases that remove damaged bases.
B) bypass DNA polymerases that can continue synthesizing DNA from a damaged DNA strand.
C) proteins that bind to deaminated bases.
D) excinuclease.
E) repair endonucleases and excinuclease.
B) bypass DNA polymerases that can continue synthesizing DNA from a damaged DNA strand
30) Which of the following is involved in proofreading during DNA replication?
A) 5' to 3' exonuclease activity of DNA polymerase
B) 3' to 5' exonuclease activity of DNA polymerase
C) 5' to 3' endonuclease activity of DNA polymerase
D) 3' to 5' endonuclease activity of DNA polymerase
E) RNA polymerase
B) 3' to 5' exonuclease activity of DNA polymerase
31) Multiple sites of DNA replication along eukaryotic chromosomes are known as
A) replicons.
B) multireplication forks.
C) helicase loaders.
D) ARS elements.
E) None of these answers are correct.
A) replicons.
32) The protein(s) that facilitate the unfolding of chromatin fibers ahead of the replication fork is
(are)
A) helicase.
B) scaffold proteins.
C) chromatin remodeling proteins.
D) replisomal proteins.
E) helicase and scaffold proteins.
C) chromatin remodeling proteins.
33) Which of the following DNA repair mechanisms is most prone to error?
A) mismatch repair
B) base excision repair
C) SOS repair
D) nucleotide excision repair
E) All DNA repair systems are equally prone to error.
C) SOS repair
34) Double-stranded break repair mechanisms
A) are error prone.
B) may join nonhomologous ends of DNA together.
C) are part of the SOS repair system.
D) may use homologous DNA to repair the damage.
E) are error prone and may join nonhomologous ends of DNA together or use homologous DNA
to repair the damage.
E) are error prone and may join nonhomologous ends of DNA together or use homologous DNA
to repair the damage.
35) Homologous recombination may result in gene conversion when used to fix double-stranded
DNA breaks because
A) sister chromatids are always identical.
B) homologous recombination is prone to errors, potentially causing permanent genetic
mutations.
C) strand invasion may introduce new genes onto the damaged chromosome.
D) homologous chromosomes may carry different alleles of the same gene.
E) the Holliday junction may migrate, resulting in loss of genetic material.
D) homologous chromosomes may carry different alleles of the same gene.
36) Transposase is
A) required to resolve a Holliday junction.
B) involved in the movement of mobile genetic elements in prokaryotic and eukaryotic genomes.
C) found only in prokaryotic transposons.
D) required for synthesis-dependent strand annealing to repair double-stranded DNA breaks.
E) necessary for replicative transposition but not conservative transposition
B) involved in the movement of mobile genetic elements in prokaryotic and eukaryotic genomes.
37) It has been observed that when plants are subjected to extreme physical stress such as
drought or high salinity, the transcription and activity of transposase genes increases. Increased
transposase activity may
A) increase the rate of mutation under stressful conditions.
B) aid in repairing DNA damage caused by stressful conditions.
C) halt DNA replication and cell division.
D) increase homologous recombination and genetic diversity.
E) explain why some plants can't withstand drought conditions.
A) increase the rate of mutation under stressful conditions.
38) Composite transposons are composed of
A) the transposase gene flanked by inverted repeat sequences.
B) a mobile gene flanked by inverted repeat sequences.
C) the transposase gene only.
D) several genes, such as antibiotic resistance genes, flanked by transposase genes.
E) several genes, including transposase, flanked by inverted repeat sequences.
E) several genes, including transposase, flanked by inverted repeat sequences.
1) A strain of mutant bacteria has been isolated. You isolate the mRNA corresponding to the mutated gene from this bacterial strain to use in an in vitro translation system and note that even in vitro it is difficult to use this message and obtain protein. Of the following, which is a likely explanation for these results?
A) There may be a mutation in the Shine–Dalgarno sequence of the DNA, resulting in mRNA that binds poorly to the ribosome.
B) There may be a mutation in the ribosomal rRNA recognizing the Shine–Dalgarno sequence of the message.
C) This mutant may have altered tRNA molecules, such that the codon–anticodon interaction during translation is affected.
D) This mutant may not manufacture enough translation factors for effective translation.
E) In this mutant, the ribosomal subunits may not associate well enough for effective translation.
A) There may be a mutation in the Shine–Dalgarno sequence of the DNA, resulting in mRNA that binds poorly to the ribosome.
2) Cells that use the amino acids selenocysteine and pyrrolysine have
A) two more codons compared to other cells.
B) two more tRNAs compared to other cells.
C) two more aminoacyl-tRNA synthetases compared to other cells.
D) two more tRNAs and aminoacyl-tRNA syntheses compared to other cells.
E) inosine at the third position of their tRNA anticodons to accommodate these amino acids.
D) two more tRNAs and aminoacyl-tRNA syntheses compared to other cells.
3) Of the following steps associated with translation, which does not involve hydrolysis of GTP? A) aminoacylation of tRNA
B) formation of the initiation complex
C) binding of the aminoacyl tRNA to the codon at the A site
D) translocation of the ribosome E) release of polypeptide
A) aminoacylation of tRNA
4) In which of the following groups are most mRNAs monocistronic?
A) bacteria
B) archaea
C) eukarya
D) bacteria and archaea
E) All mRNAs are monocistronic.
C) eukarya
5) You are attempting to determine which step of prokaryotic translation a new antibiotic inhibits. You have determined that the antibiotic interacts with a translational factor but need to identify which one. If the new antibiotic interferes with elongation, which of the following factors could be a potential site of action?
A) IF3 interaction with an aminoacyl tRNA
B) EF-Tu placement of N-formylmethionine onto the ribosome
C) EF-Tu interaction with an aminoacyl tRNA
D) EF-Ts translocation of tRNAs from the A site to the P site
E) All of these could be potential sites of action.
C) EF-Tu interaction with an aminoacyl tRNA
6) In eukaryotic initiation, one will often see the sequence ACCAUGG as a translational start sequence. This sequence is known as the ________ sequence.
A) Okazaki
B) ETS (eukaryotic translational start)
C) Kozak
D) IRES (internal ribosome entry sequence)
E) CIBS (complex initiation binding sequence)
C) Kozak
7) An E. coli strain contains a mutation in the malate dehydrogenase gene that results in a stop codon at amino acid position 14. From subcultures of this strain, a colony was obtained that was capable of making malate dehydrogenase. Your approach to examining this phenomenon should be to examine
A) the DNA sequence to see if a reversion has occurred.
B) the DNA sequence to see if a suppressor mutation has occurred.
C) other proteins to see if other stop codons are ignored, indicating the presence of a suppressor tRNA.
D) the DNA and plasmids for the incorporation of a second complete copy of the malate dehydrogenase gene.
E) All of these approaches need to be considered.
E) All of these approaches need to be considered.
8) The codon for phenylalanine is UUU. Which of the following codons also most likely encodes for phenylalanine?
A) AAA
B) CUU
C) AUG
D) AUU
E) UUC
E) UUC
38) Cytosolic polypeptides are transported to a target organelle via
A) transportin mechanisms.
B) post-translational import.
C) cotranslational import.
D) the importin protein.
E) the activity of ribosomes.
B) post-translational import.
9) During initiation of translation in both eukaryotes and prokaryotes the ________ binds before the ________.
A) large ribosomal subunit; small ribosomal subunit
B) small ribosomal subunit; large ribosomal subunit
C) EF-Tu; tRNA
D) EF-G; EF-Tu
E) ribosomes; initiation factors
B) small ribosomal subunit; large ribosomal subunit
10) In eukaryotes, the 3ʹ end of the mRNA is important in the initiation of translation because A) it contains Kozak sequences that aid in initiation.
B) translation occurs in the 3ʹ to 5ʹ direction.
C) eIF4A is attached to the 3ʹ end and removes any secondary structure that might prevent translation.
D) the 3ʹ poly(A) tail and PABP bind initiation factor eIF4G, stabilizing the 5ʹ end of the mRNA. E) it contains Kozak sequences that aid in initiation of translation in the 3ʹ to 5ʹ direction.
D) the 3ʹ poly(A) tail and PABP bind initiation factor eIF4G, stabilizing the 5ʹ end of the mRNA. E) it contains Kozak sequences that aid in initiation of translation in the 3ʹ to 5ʹ direction
11) Which of the following statements is not true concerning peptidyl transferase? A) It is a ribozyme having catalytic activity.
B) It catalyzes peptide bond formation.
C) It moves the ribosome, so translation continues.
D) It is associated with the large subunit of ribosomes.
E) It requires no outside source of additional energy, such as ATP.
C) It moves the ribosome, so translation continues.
12) Which of the following enzymes is associated with the formation of peptide bonds? A) peptidase
B) aminoacyl transferase
C) peptidyl transferase
D) peptide hydrolase
E) inteins
C) peptidyl transferase
13) Using the techniques of genetic engineering, you design a protein you want to accumulate within the ER of yeast cells. To accomplish this goal, you need to
A) do nothing; all proteins go through the ER.
B) incorporate appropriate mannose-6-phosphate groups.
C) incorporate the appropriate DNA sequence(s) to create signal sequences in the mature peptide.
D) incorporate radioactive amino acids into the protein.
E) remove the N-formyl group located at the N-terminus of the polypeptide.
C) incorporate the appropriate DNA sequence(s) to create signal sequences in the mature peptide.
14) Which of the following organelles does not receive proteins by posttranslational import of proteins synthesized on cytoplasmic ribosomes?
A) lysosome
B) nucleus
C) mitochondria
D) peroxisomes
E) chloroplast
A) lysosome
15) How do proteins become embedded in the membrane of the endoplasmic reticulum?
A) by protein chaperones that insert polypeptides into the ER membrane
B) by splicing of proteins on either side of the ER membrane
C) via stop- and start-transfer sequences that stop and start translocation of a polypeptide through the ER membrane, resulting in transmembrane proteins
D) by post-translational import can results in a transmembrane ER protein
E) via transit sequences that target polypeptides made in the cytosol for the ER membrane
C) via stop- and start-transfer sequences that stop and start translocation of a polypeptide through the ER membrane, resulting in transmembrane proteins
16) Which of the following rRNA molecules is not associated with the large subunit of the eukaryotic ribosome?
A) 28S
B) 5.8S
C) 5S
D) 16S
E) Neither 5.8S nor 5S rRNA is associated with the large subunit.
D) 16S
17) The unfolded protein response slows the production of ________ and increases the ________.
A) signal recognition particles; activity of proteasomes
B) most proteins; production of lysosomal proteins
C) protein disulfide isomerase; activity of proteasomes
D) most proteins; production of proteins required for protein folding and degradation
D) most proteins; production of proteins required for protein folding and degradation
18) Release factors of translation recognize the codon A) AUG.
B) GUA.
C) UGA.
D) GGA.
E) UUU.
C) UGA.
39) The transfer of polypeptides into the ER is called
A) transit sequence import.
B) post-translational import.
C) cotranslational import.
D) endoplasmic import.
E) ribosomal import.
C) cotranslational import.
19) Which of the following activities is not associated with post-translational processing? A) glycosylation
B) specific cleavage of polypeptides
C) chaperonin activity
D) addition of lipid groups
E) polyadenylation
E) polyadenylation
20) Ribosomes that do not anchor on the surface of the rough endoplasmic reticulum during translation most probably
A) are synthesizing cytoplasmic proteins.
B) have a defect in ribosomal proteins that allow attachment to the surface.
C) have a signal peptidase error.
D) do not make the appropriate anchor protein. E) All of the above are probable reasons.
A) are synthesizing cytoplasmic proteins.
21) AUG is the "start" codon in
A) prokaryotic translation.
B) eukaryotic translation.
C) both prokaryotic and eukaryotic translation. D) neither prokaryotic nor eukaryotic translation. E) prokaryotic modification.
C) both prokaryotic and eukaryotic translation. D) neither prokaryotic nor eukaryotic translation.
22) The initial amino acid incorporated into a nascent peptide is N-formylated in
A) eukaryotic translation.
B) prokaryotic transcription.
C) prokaryotic translation.
D) both eukaryotic and prokaryotic translation.
E) both transcription and translation in prokaryotes.
C) prokaryotic translation.