Part 2: ER translocation and Protein Maturation
1/120
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
121 Terms
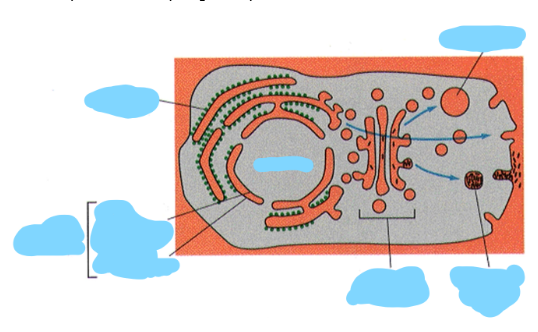
Label the parts of this topological representation of the cell
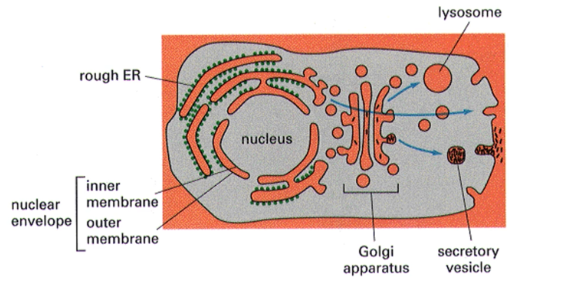
The lumen of intracellular compartments is topologically equivalent to —-
the outside of the cell
How is topology maintained during vesicle transport
as vesicles bud off from a donor compartment and fuse to a target compartment, the direction of membrane proteins always remains the same (whether they are pointing in towards the vesicle or out of the vesicle)
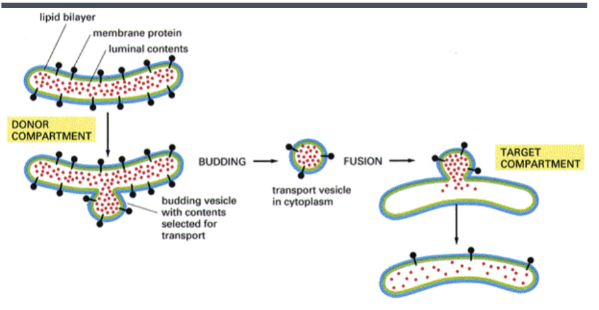
Donor compartment
compartment from which a vesicle buds off
Target compartment
compartment that a vesicle is targeted to and fuses with
General features of ER
highly convoluted single membrane-bound organelle that may contain up to 50% of cell membrane (as is the case in liver cells). The tubular nature of ER gives it a very large surface area (~25 tunes the surface area of plasma membrane in liver cells). Thought to enclose a single inner space called the lumen which can comprise >10% of the total cell volume.
Smooth ER
site of most lipid synthesis in the cell
Rough ER
site of entry of proteins into the secretory pathways. Named due to ribosome on the surface giving it a “rough” appearance
Transitional elements
part rough and part smooth area of ER. site at which transport vesicles bud off to carry newly synthesized lipids and proteins to golgi
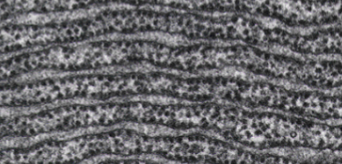
What type of ER is this?
rough
What type of ER is this?
smooth
Microsomes
small, membrane-bound vesicles formed from the fragmentation of the endoplasmic reticulum (ER) during cell disruption. They are a crucial tool in biological research because they contain enzymes involved in a wide range of cellular functions, including protein synthesis
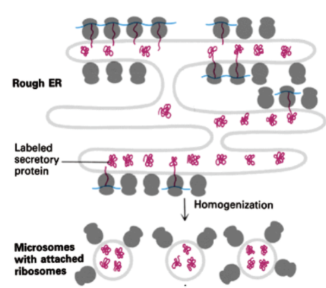
How can smooth ER and rough ER be separated experimentally?
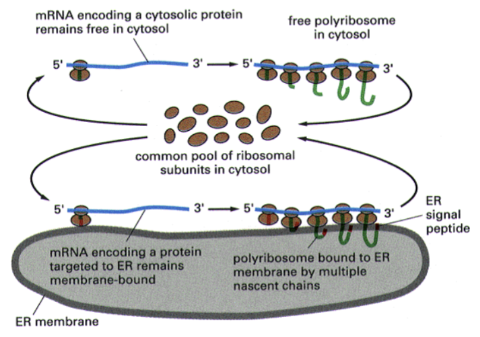
Show how common ribosomal subunits interact with mRNA encoding cytosolic proteins vs mRNA encoding a protein targeted to the ER
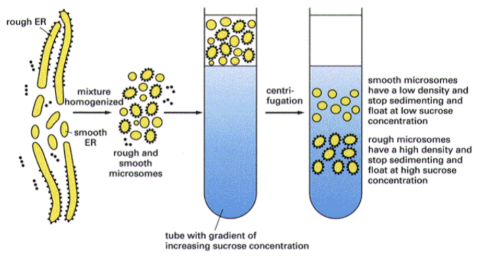
How to separate free vs membrane bound ribosomes experimentally?
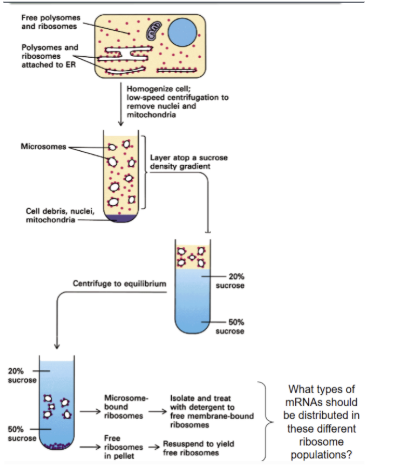
Sucrose gradient centrifugation separates molecules based on —-
density (denser materials sink further)
After separating membrane bound ribosomes and free ribosomes, what type of mRNA would be found with each?
the mRNA attached to free ribosomes code cytosolic proteins while the mRNAs attached to membrane bound ribosomes are ones that encode proteins that are targeted to a cell membrane
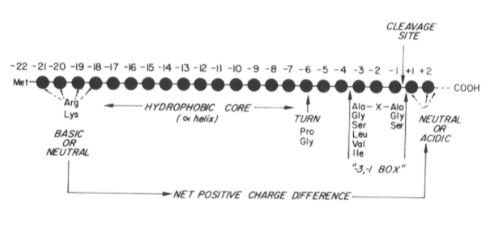
5 Key features of ER targeting signals
15-35 amino acids, a positively charged amino terminal containing R or K residues (2-10 amino acids in length). Central core of 9 or more hydrophobic or neutral amino acids that form an alpha helix. A turn inducing residue (proline or glycine) following the core regions and about 6 amino acids before the cleavage site. specific cleavage site recognized by signal peptidase and preceded in most cases by small, apolar amino acids (alanine in 50% of known examples) at positions -1 and -3 (cleavage occurs between -1 and +1)
How do peptides cross the ER membrane
signal sequence binds to the surface of lipid bilayer, insertion continues as the backbone of alpha helix straightens and pulls the polypeptide into the membrane. Basic residues at the N terminus of the signal anchors the signal to the “cis” face of the membrane (prevents headfirst insertion and creates helical hairpin/insertion loop configuration). hydrophilic proteins at then accommodated in a water-filled protein channel
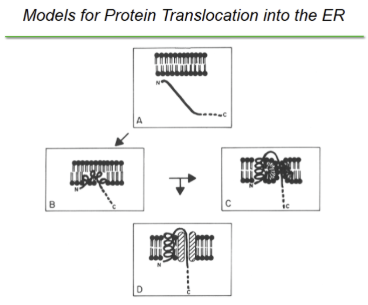
If a polypeptide is hydrophilic, in order to cross the ER membrane it must be accommodated in a —-- formed by either —- or —--. It was found experimentally that are formed by —-
water-filled channels formed by either lipids in a non-bilayer conformation or a proteinaceous channel; proteinaceous channels.
Cis face of ER
part of ER membrane facing the cytosol
Trans face of ER
part of ER membrane facing the lumen
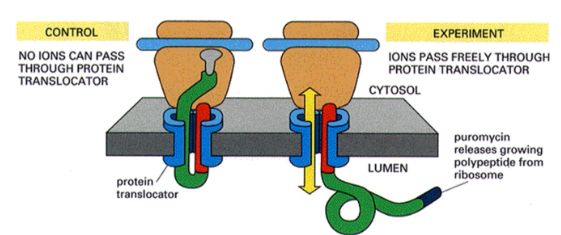
What evidence is there for a proteinaceous channel being used for transportation of peptide across the ER membrane
puromycin was added to translating microsomes to release growing peptide from the ribosome and ion currents were measured. Because ion currents were detected, it was concluded that proteinaceous channels used as a lipid line pore would just close in the event of protein release.
The original signal hypothesis
produced by Gunter Blobel. States that as protein are translated, if they have a secretory signal on the N terminus they will be targeted to the Er membrane where they will be translated through a receptor protein with associated pore into the ER lumen
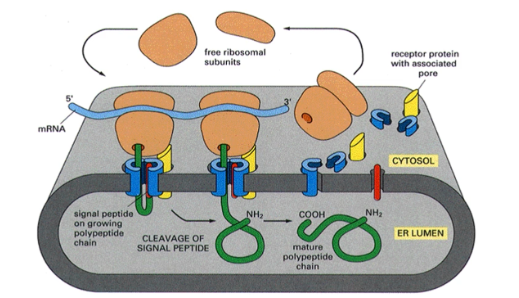
The (N- or C-) terminus is the first part of the protein to enter the ER lumen following —-
N-; cleavage of signal peptide
Two phases of translocation of proteins into the ER
ATP independent targeting of the nascent polypeptide to the cis face of the ER membrane and ATP dependent translocation of the polypeptide across the ER membrane
t/f translocation of proteins to the ER is always cotranslational
false, it can be both cotranslational and post-translational
Show the process of how a nascent protein is targeted to the ER membrane
after ~30 amino acids the nascent chain emerges from the ribosome, it is bound by the SRP and translation is arrested (occurs after ~70 AAs as 30-40 AAs are buried in ribosome. SRP complex moves to the ER and docks the SRP receptor protein complex translation arrest is relieved and SRP is released and recycled to be used again.
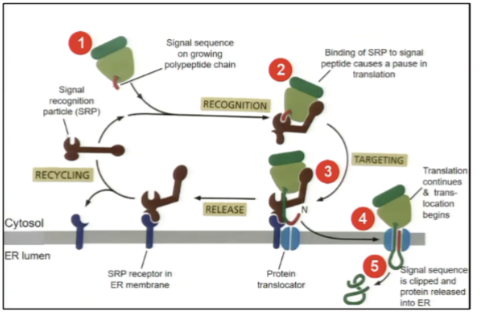
If the SRP does not dock to a SRP receptor, the ribosome may instead dock to a — on the ER membrane
ribosome receptor
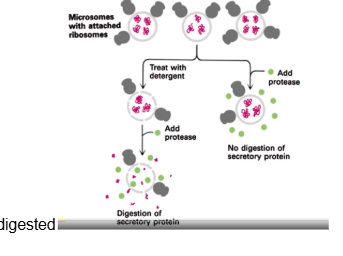
How was it proven that proteins remain in a microsome lumen when targeted there?
when you have a microsome with secretory proteins in them and protease alone is added, no digestion of secretory proteins occurs. When detergent is added to break down the microsome membrane, then protease is added the secretory protein is digested
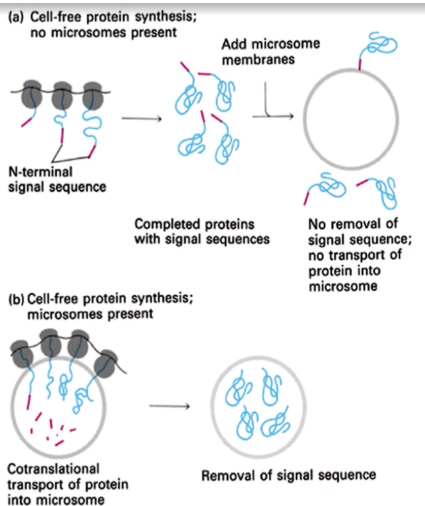
How was it proven that targeting of proteins to ER is a cotranslational process?
it was shown that if microsomes are not present during the process of translation of proteins, there would be no removal of sequence of transport of protein into the microsomes when they are added after translation
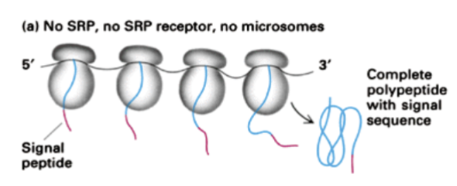
Show how translation occurs for a protein with a signal recognition sequence when no SRP, no SRP receptor and no microsomes are present
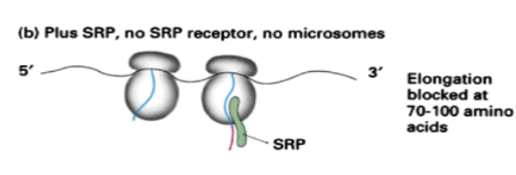
Show how translation occurs for a protein with a signal recognition sequence when SRP is present but no SRP receptor and no microsomes are present
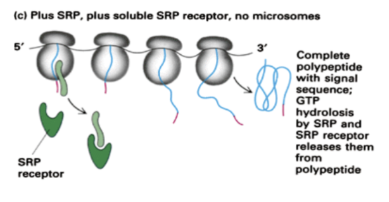
Show how translation occurs for a protein with a signal recognition sequence when SRP is present and soluble SRP receptors are present but no microsomes are present
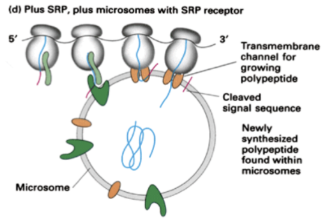
Show how translation occurs for a protein with a signal recognition sequence when SRP and microsomes with SRP receptors are present
SRP
signal recognition particle. 11s ribonucleoprotein complex composed of a 7s RNA and six polypeptide chains. In eukaryotes, Proteins are targeted to the ER by the SRP in a co-translational manner
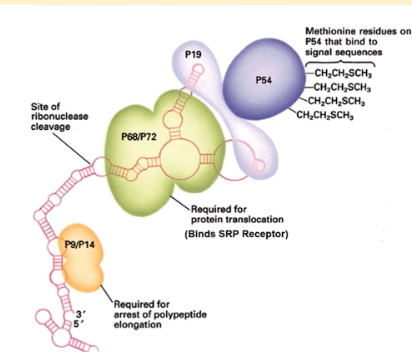
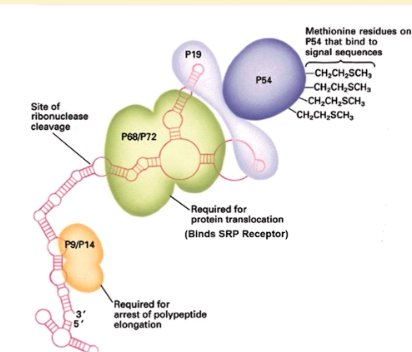
3 domains of SRP
p19/54 domain, p9/14 domain and p68/72 domain
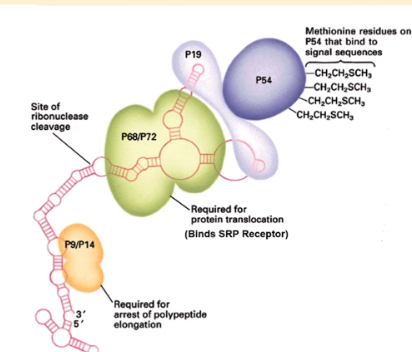
p19/54 domain of SRP
methionine residues on P54 bind to signal sequences while p19 links p54 to the 7s RNA
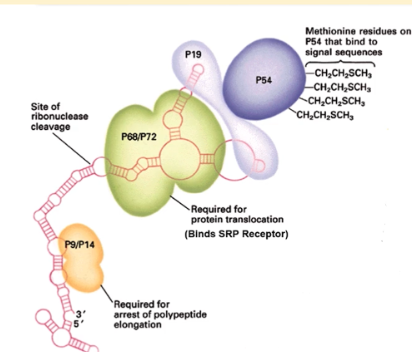
p9/14 domain of SRP
binds to the A site of of ribosome during SRP binding and arrests polypeptide elongation (critical for translation arrest)
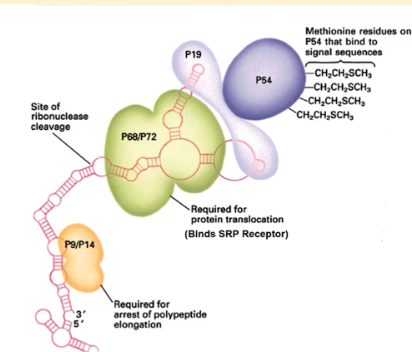
p68/72 domain of SRP
binds to the SRP receptor and initiates insertion of signal sequence into ER membrane. Critical for protein translocation
The SRP is found loosely associated with the —--, bound to both —- and —- associated ribosomes and free in the —--
RER; systolic and ER associated ribosomes; cytosol
How was the SRP originally identified?
it was found that salt-washed microsomes were unable to import secretory proteins in vitro but import may be restored when salt extract was added back
t/f (and explain why) SRP has some affinity for all ribosomes, but the affinity increases slightly upon emergence of a signal sequence during translation
false, the affinity increases 3-4 orders of magnitude upon emergence of a signal sequence
SRP receptor structure and frequency on RER surface (and what this frequency suggests)
composed of 70kDa alpha subunit and 30 kDa beta subunit. Much less abundant than ribosomes on RER surface which suggests it is not involved in subsequent stages of the process.
_—-- by the SRP receptor leads to release of SRP from the receptor so a new targeting cycle may begin
GTP hydrolysis
How does P54 of the SRP carry out proofreading?
it contains a GTP binding domains in which GTO hydrolysis may result in release of erroneously bound signal sequence from the M domain
What are the optimal conditions of a signal peptide for SRP association to occur?
before secondary and tertiary structures within the nascent chain leads to masking of the secretory signal
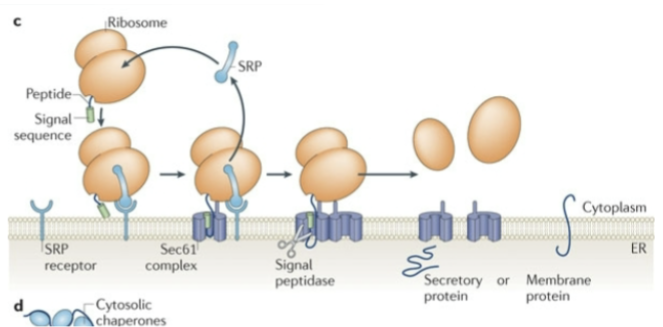
Show Co-translational translocation into the ER
Show Post-translational translocation to the ER
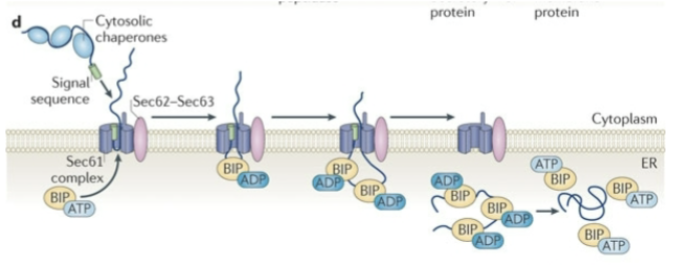
Sec62p
binds to the protein as it first associated in the ER membrane in an ATP-independent manner
Sec62p crosslinks to —-- in yeast, but only in the —-- phase of the reaction. Thus it interacts with them (earlier or later) than Sec61p
translating precursors; ATP-independent (early); earlier
Sec63p
has a lumenal J domain that associates with BiP and helps coordinate its function. This J domain is homologous to E. coli DnaJ protein which interacts with a Hsp70 homolog (e.g. BiP).
Sec61p complex
a key protein complex in the endoplasmic reticulum (ER) membrane that forms a channel for the translocation of proteins into the ER. found to interact directly with translocating polypeptides through crosslinking experiments in yeast and mammalian cells. Interacts directly with ribosomes, so it may act as a ribosome receptor
When does Sec61p crosslink to translocating precursors in yeast?
during the ATP-dependent phase of translocation
Sec61 shares structural amino acid sequence homology with the E. coli — protein
SecY
When does Sec61p crosslinking occur in crosslinking experiments?what does this indicate?
ATP-dependent phase of the transport reaction, indicating that the interaction occurs while the precursor is moving across the membrane
BIP
immunoglobulin heavy chain binding protein. Kar2p in yeast. Resident ER protein that is related to the Hsp70 class of proteins and is thought to bind aggregated or misfolded proteins to prevent them from leaving the ER. It may also help refold them using an ATP dependent activity.
BiP can be crosslinked to —-- in yeast which is consistent with studies where it was found that mutationally altered BiP blocked precursor translocation
translocating precursors
Sec62p and Sec63p are through to be particularly important for proteins that use a —---
post-translational translocation mechanism
t/f sec62p and sec63p are essential for ER translocation of all secreted proteins
false, they are not always essential
The existence of a distinct protein that functions as a —-- is controversial. The function may be carried out by Sec61p
ribosome receptor
Signal peptidase
contains 5 different proteins. The catalytic site is on the trans (lumenal) side of the ER membrane. Subunits of this complex appear to span the membrane only once and have a second hydrophobic region that may interact with signal sequences
Oligosaccharyltransferase
on the trans (lumenal) side of the ER membrane. Responsible for transfer of an oligosaccharyl moiety to an asparagine residue on a consensus recognition motif. Consists of 3 subunits (ribophorins 1 and 2 and a 48 kDa polypeptide)
t/f signal peptidase and oligosaccharytransferase are required for protein translocation
false
Minimal components for transport into the ER
Sec61 and SRP alone are sufficient for many precursors however Sec62 is required for translocation of many other precursors
Factors that influence protein topology during ER translocation
proteolytic processing, location of the signal, and charge distribution of amino acids around the signal
Polytopic vs bitopic proteins
Polytopic proteins span the lipid bilayer more than once, while bitopic proteins span it only once
Type 1 protein
transmembrane, N-terminus on trans side of membrane (lumen) [bitopic]
Type 2 protein
transmembrane, N-terminus on cis side of membrane (cytosol) [bitopic]
Type 3 protein
two or more membrane spanning segments [polytopic]
Targeting signals
regions of the protein that facilitate insertion into membranes. Generally at extreme N terminus but can be internal
Start and stop transfer sequences
stretch of 20-30 hydrophobic amino acids (also function as membrane anchor sequences)
The orientation of proteins across membranes is mediated by —- stretches of amino acids called —-- sequences
hydrophobic; start and stop transfer sequences
Polytopic proteins have —-- start and stop sequences
alternating
Start transfer sequences that follow stop sequences are —- independent
SRP
How can you determine how many membrane spanning domains a protein has?
hydropathy plot (a graph that visualizes the hydrophobic and hydrophilic regions of a protein's amino acid sequence to predict its structure). Peaks of hydrophobicity indicate transmembrane domains
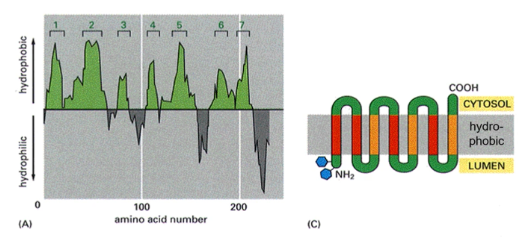
Bitopic
spans the cell membrane once
Start transfer sequences are usually preceded by —-- that act to determine the orientation of signal insertion with the N -terminus oriented toward the —--.
1 or more basic residues; cytoplasm
If the start transfer signal is followed by basic residues, the N terminus will be oriented toward the —-
ER lumen
If basic residues follow the signal sequences, the N terminus will be oriented toward what? If they precede the signal sequence, where will the N terminus be oriented?
ER lumen; cytoplasm
t/f After insertion into the ER membrane, topology of proteins will remain the same throughout the secretory pathway
true
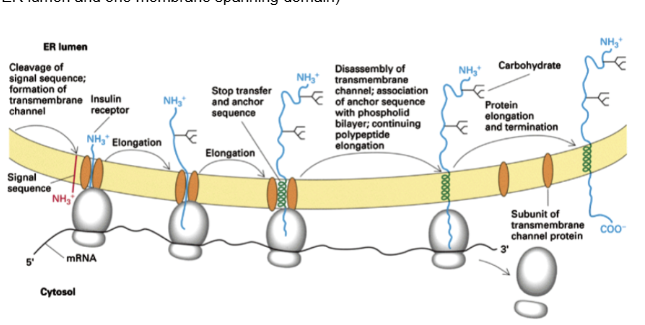
Show the membrane insertion of a Type 1 with N-terminal signal protein
(N terminus in ER lumen and one membrane spanning domain)
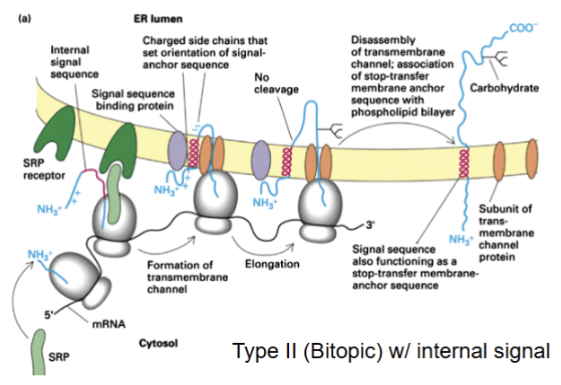
Show the membrane insertion of a Type 2 protein with internal signal
( C terminus in ER lumen and one membrane spanning domain) signal sequence as functions as stop-transfer membrane anchor sequence
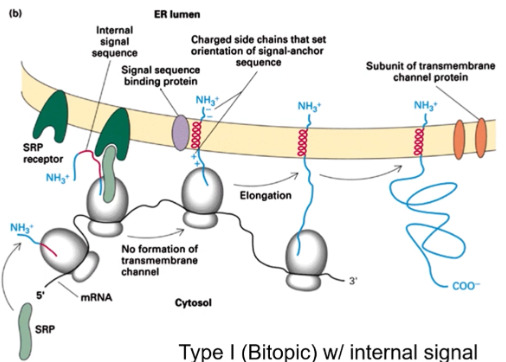
Show the membrane insertion of Type 1 protein with an internal signal
(N-terminus faces ER lumen) N-terminus faces lumen without cleavage.
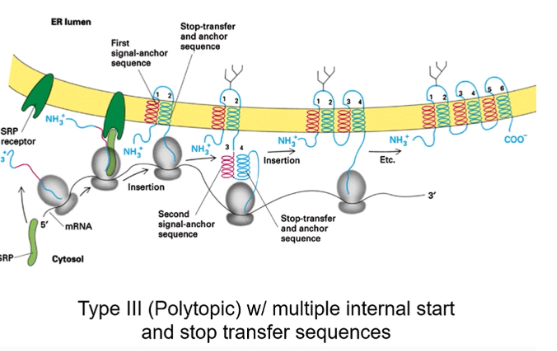
Show the membrane insertion of a Type 3 protein with multiple internal stop and start transfer sequence
polytopic with alternating signal-anchor sequence and stop-transfer and anchor sequences
Modifications that may occur during protein transport
formation of disulfide bonds, folding of polypeptide chain, additions and modification of carbohydrates, specific proteolytic cleavages, assembly of oligomeric (multimeric) proteins and quality control of proteins that are not properly folded or modified.
ERAD
ER associated degradation. If proper protein folding can’t be attained inside the ER, protein is subjected to this
Retrotranslocation
improperly folded proteins in the ER are transported from the ER via Der1 in yeast or Derlin-1 in mammals. Retrotranslocated proteins are degraded by the cytoplasmic proteosome
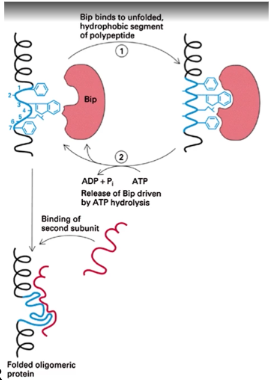
How does Bip assist in protein folding in the ER?
it repeatedly binds and releases exposed hydrophobic domains until they can form proper interactions. Also thought to bind unfolded, aggregated or misassembled proteins to prevent them from leaving the ER
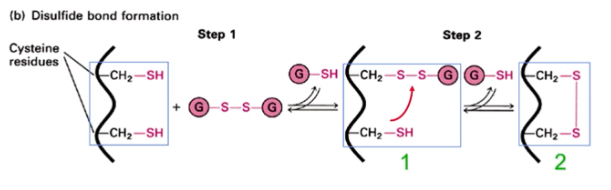
Disulfide bonds are formed in the —-- but never in the —-
lumen of the ER; cytosol
Disulfide bonds are found only in —-- proteins or in the —-- domains of —- proteins
secreted; exoplasmic domains of membrane proteins
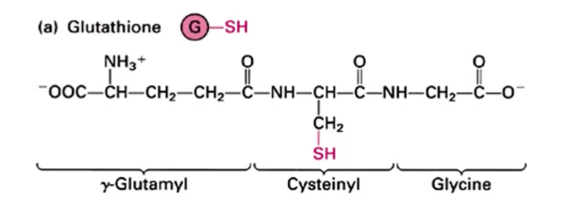
Glutathione
tripeptide present at about ~10 mM in cells as a mixture of reduced form and oxidized form. In the ER, the GSH:GSSG ratio is 5:1 which is optimal for disulfide bond formation (in the cytosol the ratio is 50:1). Prevents disulfide formation in the cytosol and catalyzes formation in the ER
What is the ratio of reduced glutathione to oxidized glutathione (GSH:GSSG) in ER? In the cytosol?
5:1 ; >50:1
Why is the ratio of GSH:GSSG higher in the cytosol than in the ER?
it is maintained by cytosolic glutathione reductase
Show the reduced form of glutathione
Show how the oxidized form of glutathione assists in disulfide bond formation
Protein disulfide isomerase (PDI)
catalyzes rapid exchange of disulfide bonds in a protein until the most thermodynamically stable configuration is found. Corrects improper disulfide bond formation
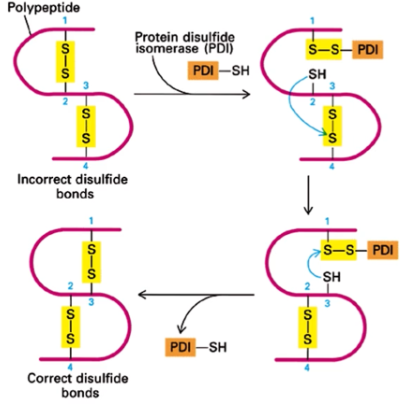
Folding and assembly of oligomeric proteins is a — process and transport from the ER is —-- until the process is complete
ordered; blocked
ER retention signals
a molecular tag, typically a short amino acid sequence, that directs proteins to remain in the ER instead of continuing through the secretory pathway. The most common signals are the KDEL sequence for soluble proteins and the KKXX sequence for transmembrane proteins
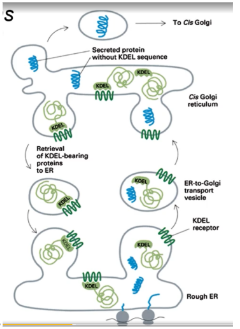
Retrograde vs Anterograde secretion
Anterograde secretion moves newly synthesized proteins and lipids from the endoplasmic reticulum (ER) to the Golgi and then to their final destination, while retrograde secretion moves materials back toward the ER from other compartments like endosomes or the Golgi