BIS2A M2 Lecture 18
1/27
Earn XP
Description and Tags
Translation - Describe the features of the genetic code • Explain the significance of reading frames • Describe the two adaptor steps for translation (tRNAs and tRNA synthetases) • Explain how a polypeptide is formed, both chemically and in the ribosome • Describe the features of ribosomes • Explain ribosome initiation, elongation and termination in prokaryotes and eukaryotes
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
28 Terms
How is a linear sequence of RNA translated into a linear sequence of amino acids?
There are 4 bases and 20 amino acids, with a combination of nucleotides. This leads to the genetic code. 4³ is 64.
Each group of 3 consecutive nuceotides (?) specifies an amino acid, or a stop to translation
codon
Explain how the genetic code is redundant but not ambiguous.
There are multiple codons that code for a single amino acid so that’s how it’s redundant. Each codon encodes for only one thing so it’s not ambiguous.
An RNA sequence can in principle be translated in 3 different reading frames. Explain this process.
There are 3 nucleotides in a codon. You can either read it at nt 1 or nt 2 or nt 3. Depending on where you start at, the rest of your codons will encode different amino acids. The cell must be able to determine the correct frame for translation initation.
What is a silent mutation
the new codon codes for the same amino acid (redundancy)
What is a missense mutation?
the new codon codes for different aminoa cids (chances the meaning/sense)
What is a nonsense mutation?
the new codon codes for a stop codon (there is no amino acid, no meaning/sense)
Translation needs some kind of adapator molecule that can linkthe nucleotide codon message with a specfiic amino acid. What does this?
tRNA - transfer
What is tRNA?
an RNA-based adaptor that binds a specific (“cognate”) amino acid on one end. It has an anti-codon on another end. This anti-codon base pairs with the codon on the mRNA, placing the amino acid in line for polymerization
What is the codon for the anticodon GAA?
a)CUU
b)UUC
c)GAA
d)TTG
GAA — CUU but because GAA is from 5’ to 3’ and CUU is 3’ to 5’, technically the codon is UUC (RNA written from 5’ to 3’)
For this transfer of amino acids to work, each tRNA must be properly attached to its amino acids. What enzyme or protein is in charge of this?
Aminoacyl-tRNA synthetases. it covalently attaches the correct amino acid to the end of each tRNA
What is the tri-partite system made of?
enzyme + tRNA + aa
What are the 2 adaptor steps of translation?
tRNA synthetase (tRNA, aa) and tRNA (aa, codon)
Protein formation via pairing of mRNA coddon and the anti-codon of an amino acid charged tRNA but how?
Chemical basis of polypeptide formation
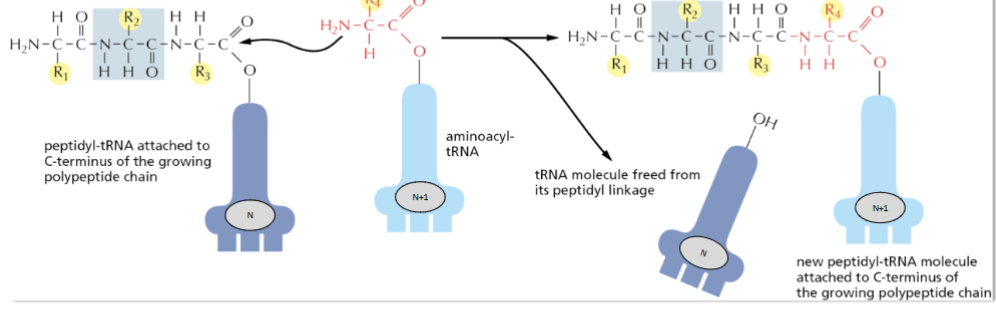
Explain what happens to the tRNA’s attached on an amino acid bonding to a peptide?
There is a tRNA attached to the C-terminus of a peptide chain. When a new amino acid bonds to the peptide chain, the tRNA(n) on the peptide chain is released. The tRNA(n+1) attached to the new amino acid stays attached to the extended polypeptide.
Where does peptide formation occur for translation?
In the ribosome which is in the cytoplasm
What are ribosomes? How do ribosomes differ from prokaryotes to eukaryotes?
Large molecular assemblies of multiple proteins and ribosomal RNA (rRNA). They are also ribozymes (RNA enzymes) that catalyze the reactions in translation. The eukaryotic ribosome is more complex but general principles are the same.
How many subunits does a ribosome have? Explain the role(s) of each subunit. Are they always joint together?
A ribosome is made of a large subunit and small subunit, each with various proteins/rRNAs. The large subunit is for peptide bond formation while the small subunit is for condon-anti-codon pairing. They come together for mRNA translation and then separate.
What provides the catalytic activity for peptide bond formation?
rRNA in the large subunit
There are 4 sites for RNA binding on the ribosome. How many sites are for mRNA and how many for tRNA?
Name the sites if applicable.
What are these sites mostly formed by?
1 mRNA binding site; 3 tRNA binding sites
A site: aminoacyl — tRNA n+1 with incoming amino acids
P site: peptidyl — tRNA n with growing chain
E site: exit — emptied tRNA n-1
They are formed by rRNA
Efficient and accurate translation is aided by?
elongation factors (GTP hydrolysis)
Polypeptide formation - explain the ribosome’s ability to proofread mistakes in tRNA’s?
the ribosome cannot break the peptide bond is the wrong tRNA (n+1) amino acid is used. But it does have initial control over the tRNA (n+1) aminoacyl using kinetic proofreading.
Why is proper initiation crucial? What are the “players” of initiation? What features of the mRNA help recruit/position the ribosome?
It sets the reading frame. (highly regulated)
The players are AUG (start codon), initiator tRNA (met), initiation factors bonded to mRNA, and ribosome subunits must come together.
MRNA features:
Euk: 5’cap, Kozak sequence
Prok: Shine-Dalgarno sequence
Kozak/S’D position the ribosome relative to the start codon
Which site does initiatior tRNA go into? A, P, or E?
P
Explain the Shine-Dalgarno sequence. What is it unique to?
A ribosome binding/recruitment sequence in prokaryotes. It is upstream of the start site. It recruits ribosomes, positions it close to the start site. Ribosome will first bind SD, then start “scanning” for the start site.
How does the ribosome recognize the Shine-Dalgarno sequence?
Base-pairing interactions
Explain how translation initiation in prokaryotes works. (4 steps)
Initiator tRNA-Met is loaded in P site of the small subunit
Small subunit is recruited to mRNA and scans for Shine-Dalgarno Sequence.
Once Sd is found, the large subunit joins the small subunit
Additional proteins (initiation factors) are involved.
Recall that mRNA is polycistronic (encodes for multiple proteins). Is there only 1 SD sequence in mRNA or are there multiple?
There are multiple SD sequences to allow for translation of polycistronic mRNAs. Ribosomes are recruited to each SD and must know where to start and where to stop. Proteins on the operon might be encoded on different reading frames. (Note: Lack of 5’ cap)