structural dna nanotech exam 2
1/68
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
69 Terms
Robust vs conventional nanostructures
robust: maintains structure and function in biological settings
conventional: can degrade or disassemble in challenging environments
Assembly of robust nanostructures vs conventional nanostructures
robust: monovalent solns, which mimic physiological conditions, can be used in vivo
conventional: high concentrations of divalent ions (not present in most biological systems), used in vitro or ex vivo
Describe the goal of 3D self-assembly of robust DNA motifs and some characteristics
-organize other species, ranging from nanoelectronics to potential drug targets.
-does not require ultra-high-resolution crystals, must be very well organized (~3 Å or better) for this form of crystallization to be of use
Two different principles of multi-state nanomechanical devices
[1] DNA structural transitions (can be based on contents of solution or by forming knots)
[2] programmed sequence-dependent devices whose states can be addressed or created individually
Characteristics of robust devices
behaves like a macroscopic device made of simple machines:
[1] a lever is either up or down
[2] a torsional element is either rotated or not
[3] a movement occurs or it doesn’t
Definition of a robust device
all of the molecules undergo the same transition, looking the same after the transition as they did before
Definition of a non-robust device
if strands of its framework can recombine and perhaps form another species OR if exchange of strands occurs while in an intermediate state
Shape-shifter devices
molecules that have different structures under different conditions, something must upset equilibrium of the molecule for it to change shape
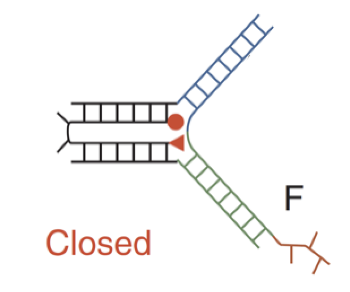
Closed sequence-dependent tweezer
when F binds to the rest of the device, it brings the two double helical domains together; red section of F aka toehold bind to complementary toehold of F’ which forms a F-F’ duplex (waste) and restores the open state
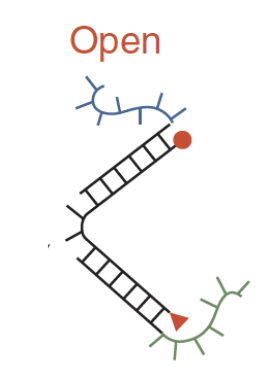
Open sequence-dependent tweezer
dyes are too far apart to interact; they are brought together by a strand F, where part of F is complementary to the blue unpaired section and part of F is complementary to the green unpaired section
PX-JX2 device
based on the PX state of DNA and one of its topoisomers, the JX2 state; makes tweezer system robust by preventing dimerization during transitions
1a. What are the 2 components of 2D and 3D DNA origami constructs?
The two key components of 2D and 3D DNA origami constructs are:
A long single-stranded scaffold strand — typically derived from M13 bacteriophage DNA, which provides the continuous framework that folds into the desired shape.
Multiple short staple strands — synthetic oligonucleotides that bind to specific regions of the scaffold to “staple” it into the programmed 2D or 3D structure through complementary base pairing
1a. What type of crossover is used to fold a DNA origami structure?
DAO
1a.Describe the difference between the components of an RNA origami construct and a DNA origami construct
DNA origami uses many short staple strands to fold a long DNA scaffold into shape.
A single continuous RNA strand that self-folds into the target shape through intramolecular base pairing and secondary structures (hairpins, junctions, pseudoknots).
No separate “staples” — folding is encoded entirely in the RNA sequence
1a. How was RNA origami generated in vivo?
The entire RNA origami design is encoded as a DNA gene construct and inserted into a plasmid vector.This plasmid is then transformed into E. coli. Inside the cell, the cell’s own RNA polymerase transcribes the designed RNA sequence.
As the RNA strand emerges from the polymerase, it folds co-transcriptionally into the intended 2D or 3D origami shape via intramolecular base pairing and structural motifs (hairpins, kissing loops, junctions)
No staples or external folding agents are needed — the structure forms autonomousl
1b. What is the difference between a DNA origami tile and a DNA brick?
DNA origami tiles are scaffold-based structures folded by short staple strands, whereas DNA bricks are scaffold-free, modular units that self-assemble through complementary domain pairing into arbitrary 2D or 3D shape
1b. What is the difference in the constitutional strands between a 2D DNA brick and a 3D DNA brick?
2D DNA brick = strand is 42-mer long strand segmented in 4 regions of either 10 or 11 bases
3D DNA brick = strand is 32-mer long strand segmented in 4 regions of 8 base
2a. What type of DNA tiles were used for the XOR computation experiment in 1D? in 2D?
The 1D cumulative XOR used TX tiles.
2D XOR computation: The 2D version used a set of seven Wang-type tiles to produce algorithmic assemblies such as the Sierpiński triangle
2a. Where was the input and output signals encoded in the tiles?
The bottom domains of the calculational tiles carried the input signals, represented by sticky-end sequences corresponding to logical 0 and 1.
The output signal was encoded in the upper sticky end of each XOR tile.
As in 1D, the inputs were encoded in the sticky ends on the bottom or lateral edges of each XOR tile.
The output was represented on the top edge, where new tiles could attach based on the computed value (0 or 1), thereby propagating the pattern upward through the 2D lattice.
2b. What is the difference in topology between the reporter strand generated for the XOR computation experiment in 1D and the one generated for the 3-colorability graph?
XOR reporter = linear (open chain); 3-colorability reporter = cyclic (closed loop
2b. How do Wang tiles assemble?
Wang tiles assemble by following a local matching rule: each tile can attach to another only when the “colors” (or sequences) of their adjacent edges match.
3a. What are the 2 ways to change the color of a crystal with a triplex strand?
1. pH-dependent TFO binding and release (acidic = colored; neutral = clear).
2. Toehold-mediated strand invasion and displacement (competitive replacement of dye-labeled TFOs)
3a. Where are Cy3 and Cy5 dyes usually attached to a DNA strand?
5′ End Labeling: The dye is covalently linked to the 5′ phosphate group via a C6 or C7 carbon linker.
3′ End Labeling: The dye is attached to the 3′ hydroxyl group of the terminal nucleotide.
Internal Labeling : Cy3 or Cy5 can be incorporated at internal modified bases (e.g., amino-modified dT or dC
3b. Describe one way to change the color of a DNA tensegrity triangle self-assembled crystal that has only a single molecule in the crystallographic repeat at neutral pH.
The color of a single-molecule DNA tensegrity triangle crystal at neutral pH can be changed by toehold-mediated strand displacement, in which a fluorescently labeled strand within the crystal lattice is replaced with another strand carrying a different dye, altering the crystal’s color without disrupting its structure
3b. List 2 ways of making purple crystals.
1. Labeling DNA triangles with either Cy3 and Cy5 dyes
2. Adding a Cy3-labeled TFO strand to a triangle with a Cy5-labeled central strand
4a. In the assembly line experiment, what were the characteristics of the walker?
The walker is a triangle with 3 arms and 4 feet.
In the assembly line experiment, which state was the off state and which state was the on state for the cassettes?
OFF state = JX2 state
ON state = PX state
4b. In the assembly line experiment, what were the 3 cargos that the walker could carry?
One 5-nm gold particle (A)
Two 5-nm gold particles (B)
One 10-nm gold particles ©
4b. In the assembly line experiment, what are the eight different products possible in the assembly line? (2.5 pts)
no cargo, A only, B only, C only, A+B, A+C, B+C, and A+B+C
5a. In Adleman’s Hamiltonian Path experiment, what enzymatic techniques were used to connect the cities covalently together?
Kination + ligation
5a. In Adleman’s Hamiltonian Path experiment, why are there no phosphates on the routes? What role do the routes play in the reaction?
Phosphates are used to enable strands to be ligated to another strand, routes should act as catalysts to promote ligation of the two cities connecting together
5b. In Adleman’s Hamiltonian Path experiment, what was the purpose of the PCR reaction step?
DNA polymerase was used later to amplify the correct-length paths representing potential Hamiltonian paths (those that started and ended with the correct cities)
5b. In Adleman’s Hamiltonian Path experiment, what technique was used to assess the correct sequence of the Hamiltonian path?
DNA sequencing
6a. What is the constituent of a PNA backbone?
A PNA (peptide nucleic acid) backbone is composed of repeating N-(2-aminoethyl)-glycine units linked by peptide bonds instead of the sugar–phosphate backbone found in DNA or RNA.
In other words, the constituent of a PNA backbone is a pseudopeptide chain made of N-(2- aminoethyl)-glycine monomers, to which the nucleobases (A, T, G, C) are attached via methylene carbonyl linkers.
6a. What type of DNA motifs was used to estimate the helicity of a hybrid PNA:DNA duplex in 2D?
DX and DX+2J to form an AB* array
6a. What technique was used to estimate the helicity in 2D?
AFM
6b. What type of DNA motif was used to estimate the helicity of an exotic XNA:XNA duplex in 3D?
2-turn DNA tensegrity triangle
6b. How was the correct helicity visually assessed?
Observation of crystals formation under light microscope
7a. What is the major problem that can cause errors in the motif capture system and in algorithmic assembly?
The formation of kinetic products by the competition of half-right tiles with completely right tile.
7a. What technique was used to open the 3D DNA origami box?
The 3D DNA origami box was opened by toehold-mediated strand displacement, triggered by DNA “keys” that unlock the box’s lid.
7b. In the self-replication of BX tiles, why was the chosen repeating pattern ABBABAB instead of ABABABA?
The pattern ABABABA is palindromic — it reads the same forward and backward.
This symmetry would make it impossible to distinguish which end of the strand is the “start” or “end”
As a result, the complementary daughter could assemble in either orientation, leading to ambiguous templating.
7b. In the self-replication of BX tiles, what is the role of the perpendicular sticky-ends of the 1st generation tiles?
Each BTX tile has two sets of sticky ends:
Horizontal sticky ends → connect A and B tiles side-by-side within a generation (forming the original pattern).
Perpendicular sticky ends → project from the array.
After the parent array is formed, the perpendicular sticky ends serve as binding sites for complementary tiles of the next generation.
These perpendicular interactions ensure that the daughter array forms attached and parallel to the parent — a template-directed replication process.
Once the daughter array is complete, the two layers can be separated, leaving the newly replicated pattern intact
8a. What are the 2 types of functional groups combination that react to make nylon?
(1) Amine group (–NH₂)
(2) Either Carboxylic acid (–COOH) or an acid chloride (–COCl)
8a. Where are the functional groups attached on the DNA backbone?
2’ carbon on the sugar
8b. What triangular system was used to position PANI and PPV in 3D?
Fused AB system
8b. What modification was done to the triangular system to precisely localize the PANI and PPV?
The central strands of the triangles were extended.
8b. How were HPV-DNA and OANI-DNA conjugates templated by a DNA origami scaffold?
Specific staple strands in the DNA origami were extended with single-stranded overhangs (“handles”) complementary to the DNA sequences attached to the HPV–DNA and OANI–DNA conjugates.
These handles served as binding sites, ensuring that each polymer–DNA conjugate hybridized only at its designated position on the origami scaffold.
9a. In the 3-state device, what kind of operation transforms the device from the PX state to the JX2 state?
Rotation
9a. In the 3-state device, what kind of operation transforms the device from the JX2 state to the BX state?
Translation
9a. In the 3-state device, what kind of operation transforms the device from the BX state to the PX state?
Screw rotation
9b. To demonstrate the robustness of DNA mechanical device controlled by hybridization topology, what were the 2 experimental techniques used to the demonstrate the operation of the PX-JX2 device?
1st technique: Native PAGE
2nd technique: AFM
9b. How are the markers arranged in the PX state? in the JX2 state?
PX state: triangles are lined up on the same side
JX2 state: zigzag arrangement
10a. Where are metals like silver and mercury positioned in the DNA duplex?
Metals like silver (Ag⁺) and mercury (Hg²⁺) are positioned between mismatched base pairs within the DNA duplex, where they act as coordination bridges that stabilize otherwise unstable pairings.
Ag⁺ ions typically coordinate between cytosine–cytosine (C–C) mismatches, forming a C–Ag⁺–C metal-mediated base pair.
Hg²⁺ ions typically bridge thymine–thymine (T–T) mismatches, forming a T–Hg²⁺–T pair.
10a. Why do carbon nanotubes need to be functionalized to be positioned on a DNA origami substrate?
Carbon nanotubes must be functionalized (typically with DNA strands or reactive chemical groups) so that they become water-dispersible and can specifically hybridize to complementary sites on a DNA origami substrate, allowing precise nanoscale positioning.
10b. List 2 imaging techniques used to visualize gold nanoparticles on 2D DNA arrays.
1st technique: Atomic Force Microscopy (AFM)
2nd technique: Transmission Electron Microscopy (TEM)
10b. What imaging technique was used to visualize the 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron?
Cryo-electron microscopy
What crossovers are present in DNA origami?
Periodic and scaffold
How is DNA origami made?
ssDNA + staple strands + thermal annealing = origami
What are the four steps of the Hamiltonian exp?
[1] ligate all possible paths w T4 ligase
[2] size-select 140mers with a gel
[3] PCR-select only Quito-Shanghai strands
[4] affinity-select strands w each intermediate, then sequence
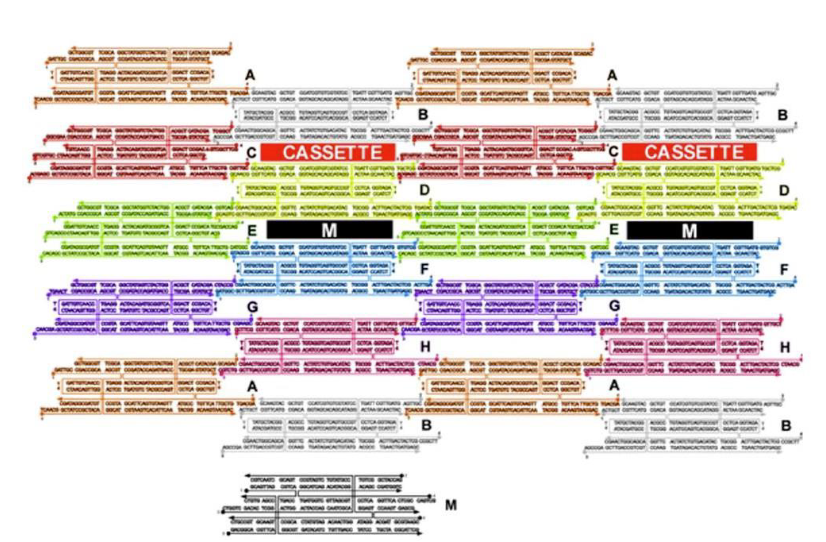
What type of DNA motif has been used to build the 2D array?
TX motif
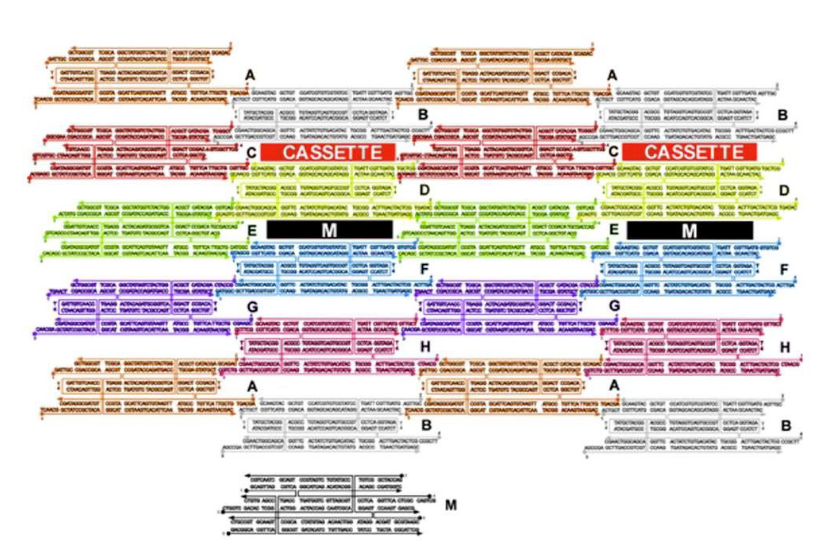
How are the cassettes oriented in the array?
Perpendicular to the array between TX tiles
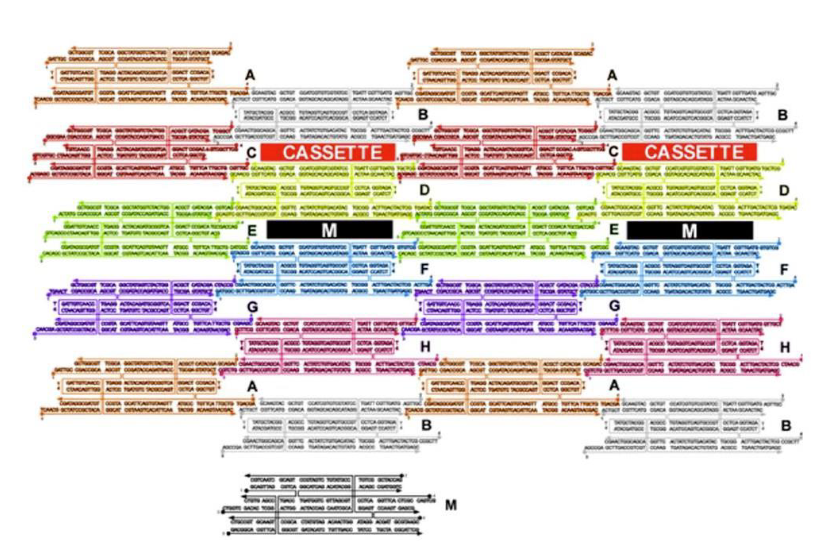
What is being represent in black (labeled M)?
Marker
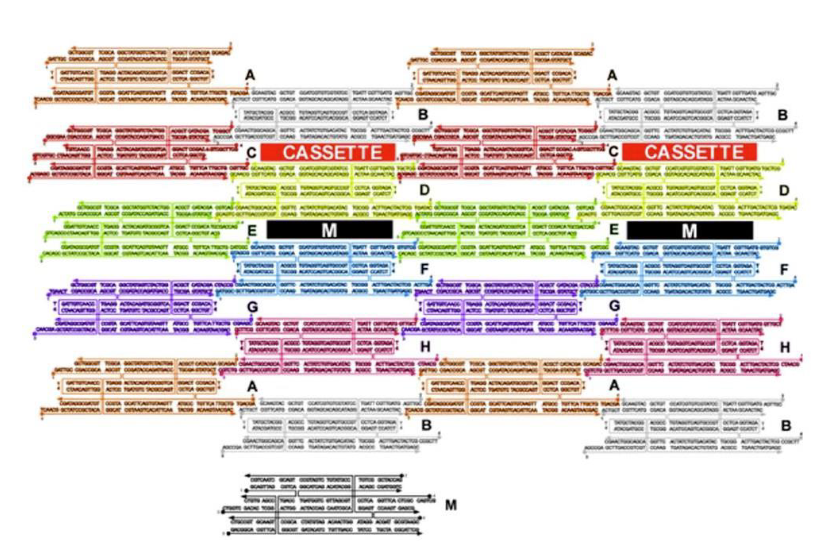
What type of DNA motif makes up tile M?
TX motif
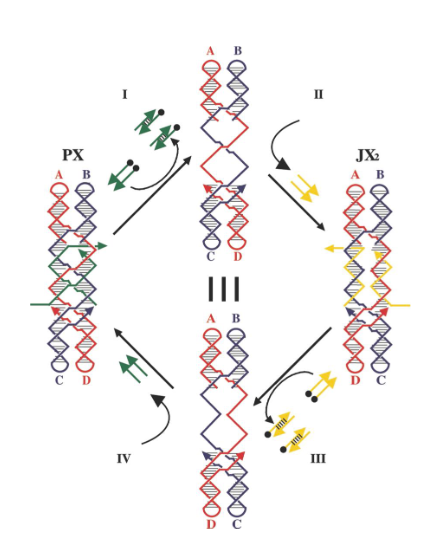
What kind of strands (in green) maintain the device in the PX state in the left?
Green set strands
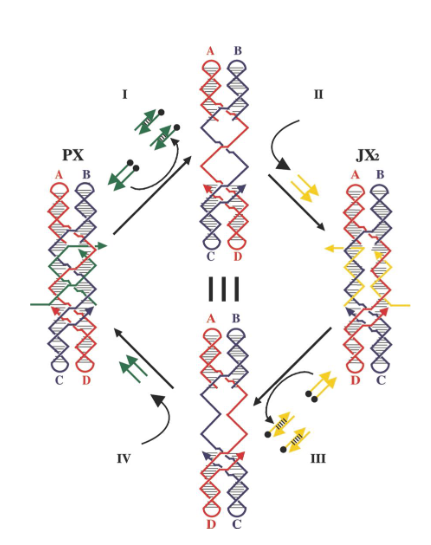
What do single strands extensions on the green strands represent?
Toehold
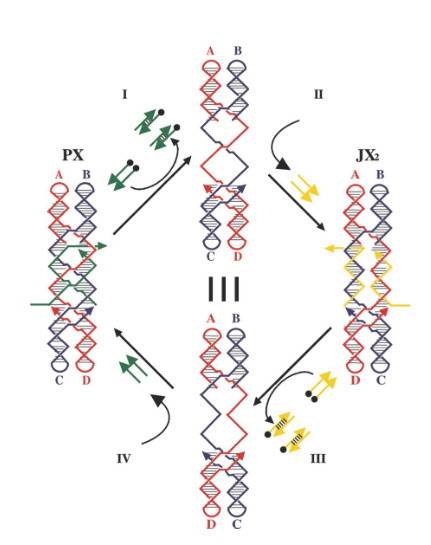
What kind of strands are added to remove the green strands?
Fuel strands
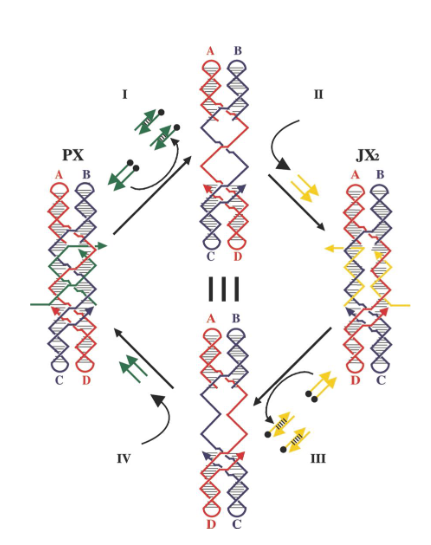
What is represented by the black dots on the green strands and what are their roles?
Biotin groups that get removed by addition of streptavidin
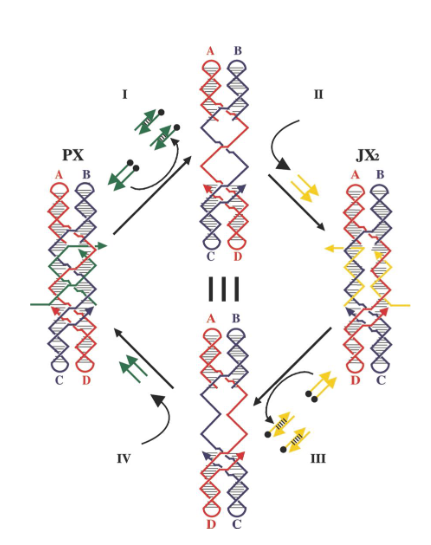
What happens when the yellow strands are added?
They set the device to the JX2 state
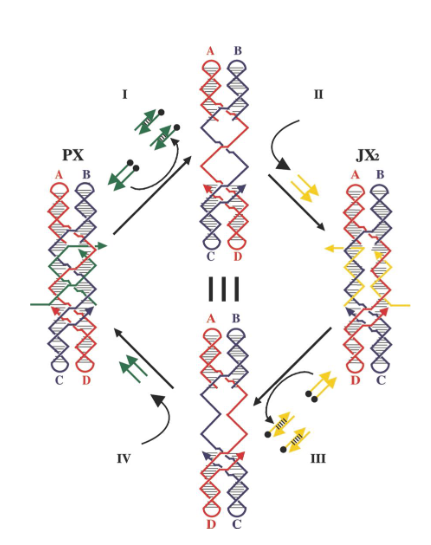
How are the yellow strands removed?
By addition of yellow fuel strands