Lecture 10 - Ribosomes and Translation
1/41
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
42 Terms
tRNA
first sequence was reported in 1965
73-93 nucleotides, synthesized by RNAP III with type 2 internal promoters
final molecules are highly processed from primary RNA transcripts (requires successive cleavage and ligation reactions, many unusual base mods)
yeast has 272 nuclear tRNA genes, 59 are interrupted (single intron)
no consensus sequence to be recognized by splicing enzymes
splicing depends on recognition of common secondary structure in RNA
plants amphibians, animals also have interrupted tRNA genes
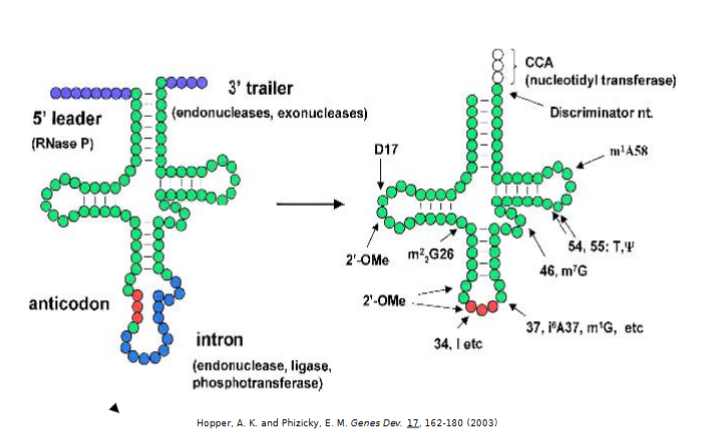
tRNA Modifications after Synthesis
D- dihydrouridine
i- inosine
T- thymidine
Ψ - pseudouridine
m - methyl group
are essential for stability, function, and structure of tRNA
tRNA Synthesis and Splicing
are heavily modified with 1% of the yeast genome codes for factors involved in tRNA processing and mod
5’ leader sequence removed by RNase P (highly conserved)
3’ trailer sequence removed by endonuclease and exonuclease
CCA added to the 3’ end by nucleotidyl transferase
intron is spliced out by multiple enzymes
additional modifications at multiple residues (all bases can be modified by other enzymes)
Bacterial vs. Eukaryotic RNase P
bacterial - ribozyme
eukaryotic - nucleolar RNP enzyme
Conversion of mRNA into a Protein
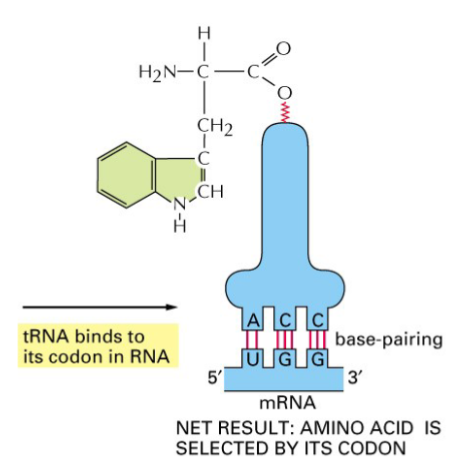
tRNA is linked to a particular aa and it recognizes a codon in mRNA so that the corresponding aa can be added
amino acids may have multiple codons but one tRNA has ONE unique tRNA
plants and animals have more tRNAs than bacteria - several different tRNAs for most amino acids
each tRNA is recognized by 1 of 20 enzymes - aminoacyl synthetase is first level of specificity in amino acid choice (one enzyme will attach one of 20 aa-charging tRNA)
after amino acid is attached by charged tRNA, tRNA can recognize codon in mRNA and bring to growing polypeptide chain
anticodon: part of tRNA that does specific interaction with codon in mRNA, a stretch of 3 sequential nucleotides
tRNA function is dependent on 3D structure
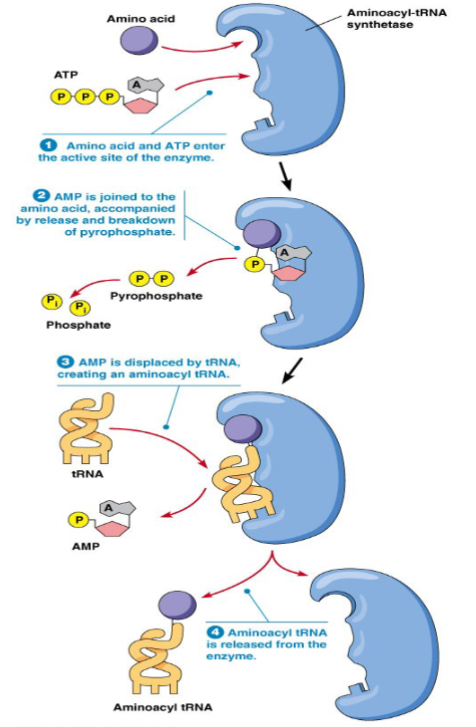
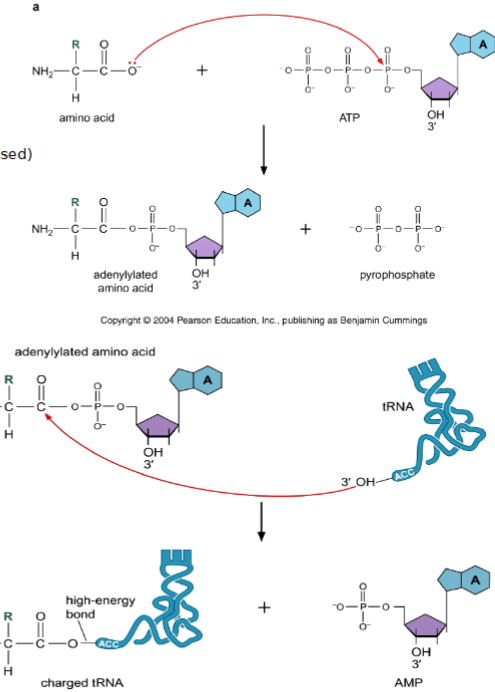
tRNA Charging
aminoacyl-tRNA synthetase (ARS) catalyzes attachment of amino acid to free 2’ or 3’ OH of the ribose of the adenosine in the 3’ end of tRNA
amino acid and adenosine monophosphate (AMP) are combined on ARS
transferring of aminoacyl group from the enzyme complex to the tRNA ‘activates’ the amino acid residue → charged tRNA
there are different enzymes based on if amino acid is added to 2’ or 3’ OH
20 different synthetases (one per amino acid)
ribosome recognizes tRNA NOT the carrying amino acid (fidelity of the aminoacyl tRNA synthetase is crucial
Which part of the tRNA is the acceptor stem?
at the 3’ end where CCA is added as a step in tRNA processing
Steps of tRNA Charging
adenylylation of amino acid (uses ATP and releases inorganic phosphate PPi)
transfer of the adenylylated amino acid to 2’ or 3’ OH of the ribose of the A on (CCA at the 3’ end of tRNA) - charged tRNA
Non-Standard Base Pairing
cells have around 50 tRNAs for 20 amino acids
single tRNA can recognize more than one codon corresponding to an amino acid due to non-standard pairing in ‘wobble positions’
antiparallel interaction - codon of mRNA is 5’-3’ and anticodon of tRNA is 3’-5’
1st nucleotide from codon is recognized by 3rd nucleotide from anticodon and vice versa
Great variability in recognition of ____ nucleotide in mRNA by ___ position in tRNA.
3rd - wobble nucleotide; 1st - wobble position
Aminoacyl tRNA Synthetase
ARS for arginine, leucine, and serine have to recognize 6 different tRNAs with different anticodons
has to select the proper amino acid to attach to the tRNA using the anticodon as the determinant
Ribosome
complex of rRNAs and proteins that directs elongation of a polypeptide (protein) - 2 subunits
synthesizes 17-21 aa/sec in prokaryotes, 6-9 amino acids/sec in eukaryotes
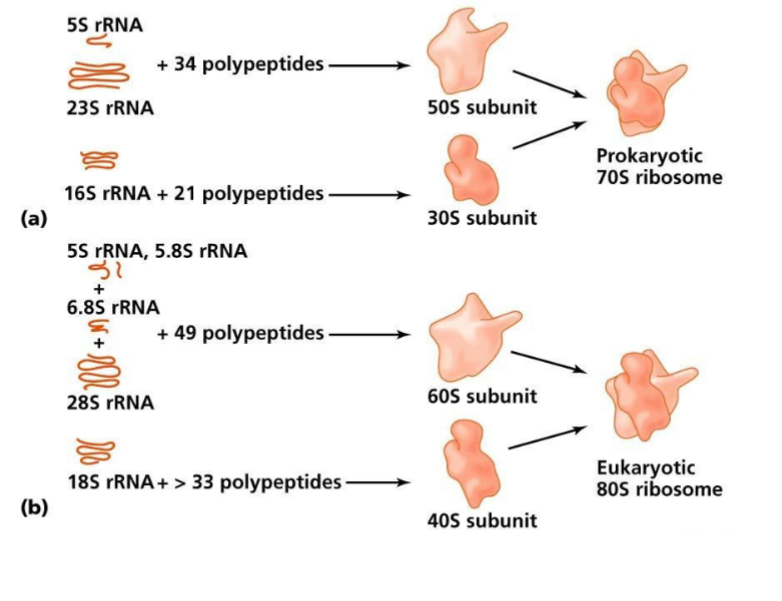
subunits of ribosome are designated in Svedburg units (S), named after inventor of ultracentrifuge
revealed differences between prokaryotes and eukaryotes’ ribosomes
What is S in Svedburg units?
a measure of sedimentation rate (velocity) of suspended particles when centrifuged under constant conditions - depends on size and shape of the particle
good measure of relative size is one is comparing same types of molecules (bigger S = faster sedimentation velocity)
Prokaryotic vs. Eukaryotic Ribosome
both have 5S rRNA subunit
prokaryotes have 16S rRNA subunit vs. 18S rRNA in eukaryotes
50S + 30S → prokaryotic 70S ribosome
60S + 40S → eukaryotic 80S ribosome
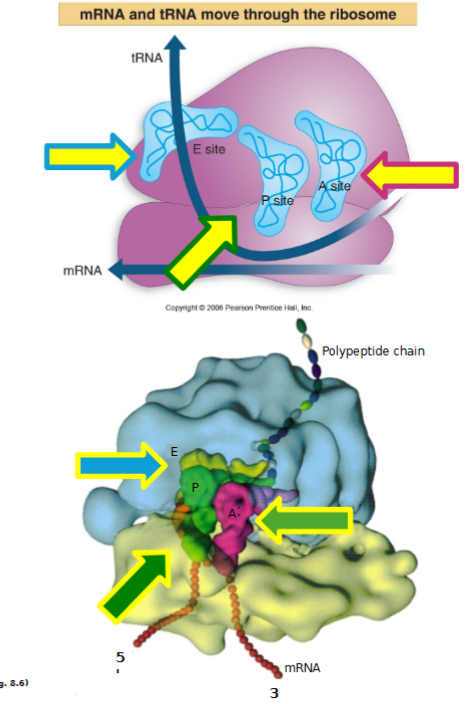
3 tRNA Binding Sites
assembled ribosome has APE sites
aminoacyl site: incoming aminoacylated tRNAs bind
peptidyl site: growing polypeptide chain is usually here where peptydyl-tRNA is
exit side: exiting (empty) tRNA site
each site is located in the cleft of the small subunit, and has adjacent mRNA codons that are translated into amino acids
Stages of Translation
3 stages:
initiation: assembly of a complete ribosome on an mRNA molecule at a correct point
elongation: repeated cycles of amino acid addition
termination: release of the new protein chain
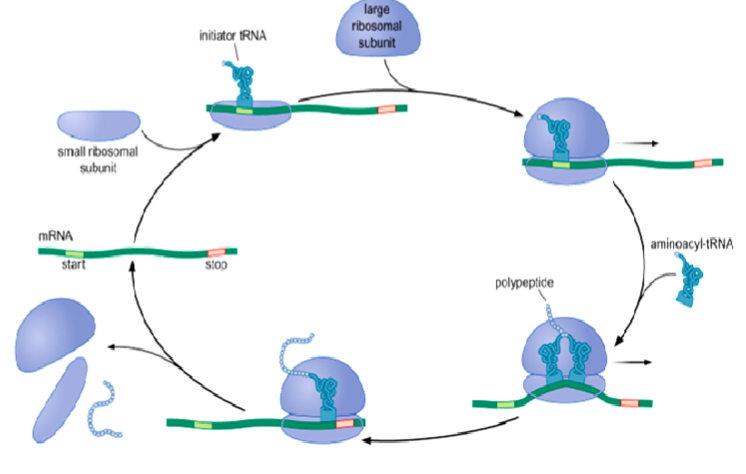
Initiation
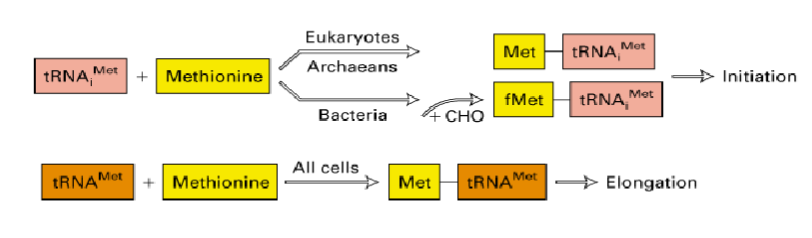
protein synthesis starts with initiator tRNA correctly positioned at start codon (AUG - Met)
euks and proks have 2 types of Met tRNAs charged with the same enzyme - methionyl tRNA synthetase
tRNAMet is for methionine at internal codons of growing polypeptide (internal AUGs)
tRNA iMet is for initiation where bacteria have modification-formyl-methionine
initiator tRNA allows for more regulation of initiation - alternate starts need to be recognized in bacteria
initial Met often removed during or immediately following translation
Prokaryotic Initiation of Translation
there is a conserved sequence in mRNA 8-13 nucleotides upstream from the first codon to be translated - ribosome binding site
Shine-Delgarno 5’ - AGGAGGU - 3’
it base pairs with complementary sequence at 3’ end of 16S rRNA in the small 30S subunit that interacts with Shine-Delgarno
positions the ribosome correctly upstream from the initiation codon
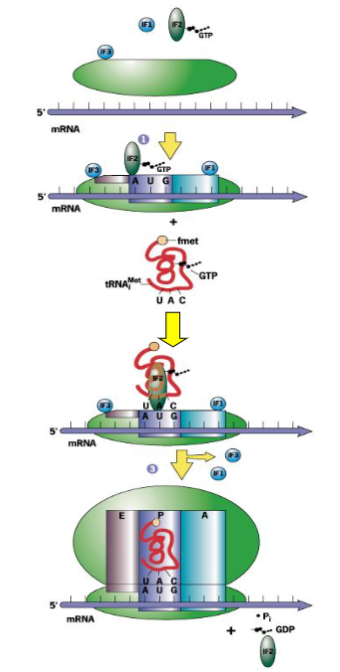
Steps of Prokaryotic Translation
IF3 binds to free 30S subunit to prevent 50S subunit from binding
IF1 binds to prevent potential binding of tRNA to A site (amino acid landing)
IF2 is a GTPase that complexes with GTP and binds
mRNA binds to 30S through interaction of Shine-Dalgarno sequence with 16S rRNA by complementary bps
initiator tRNA binds by codon-anticodon base pairing to P site
this is the 30 initiation complex
50S subunit binds
this displaces IF1 and IF3; GTP is hydrolyzed and IF2 is released
this is 70S initiation complex - ready to begin elongation
Eukaryotic Initiation of Translation
40S ribosomal subunit first binds initiator tRNA then 40S subunit-initiator tRNA complex binds mRNA and scans along mRNA until it reaches the right AUG and positions initiator tRNA there
first AUG ha to be in correct context where the optimal sequence context is Kozak consensus sequence
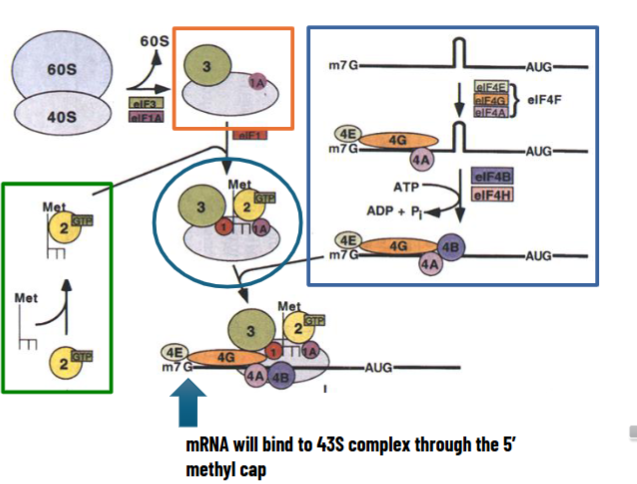
Steps of Eukaryotic Initiation
free 40S subunit complexes with eIF3 (large protein) and eIF1A to keep it apart from 60S subunit
ternary complex forms: initiator tRNA, eIF2 (GTPase), and GTP
ternary complex binds with 40S along with eIF1 → 43S preinitiation complex
on the other side, different eIF4 factors are involved in recognition of 5’ methyl cap and keep mRNA free of secondary structures using ATP E
mRNA will bind to 43S complex through 5’ methyl cap
43S complex scans mRNA to find the right AUG inside the Kozak sequence by ATP hydrolysis energy
when Kozak sequence is found, eIF5 binds and displaces ALL other factors including eIF-GDP → 40S initiation complex is formed
60S subunit binds by GTP hydrolysis
80S initiation complex is formed
eIF2-GDP complex is recycled by ______ and converted back into ____ and ____.
eIF2B; eIF2 and GDP
GTP hydrolysis happens before large subunit binds in both prokaryotes and eukaryotes
Control of Beginning of Translation
eukaryotic cells have evolved elaborate mechanisms for translational control because of:
changes in nutrient availability (mostly amino acids)
changes in available cellular energy
response to various forms of stress
hormones and growth factor stimuli
control through:
phosphorylation of translation factors
multiple AUG codons in 5’ UTR
internal ribosome entry site (IRES)
where multiple AUGs and IRES frequently act together
In some mRNAs, inhibitory secondary structure in the ___ ____ impair efficient scanning of the ___ ____ ___ for the AUG start codon.
5’ untranslated region (UTR); small ribosomal subunit
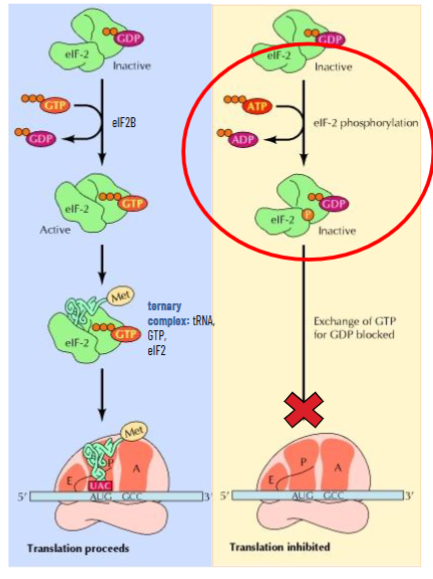
Phosphorylation of Translation Factors
eIF2 is usually recycled by eIF2B with GTP and makes the ternary complex with tRNAMet BUT translation initiation of some genes can be down-regulated by phosphorylation of eIF2
amino acid starvation (and others) leads to accumulation of uncharged tRNAs
this activated protein kinase that will phosphorylate eIF2 to make it inactive
phosphorylated eIF2 cannot be recycled by eIF2B
this means when amino acid level is low, translation of some genes will slow or shut down
Multiple AUG Codons
some viral mRNA that use host translational machinery don’t have 5’ methyl caps but they have very long 5’ UTRs
multiple AUGs in their 5’ UTRs (also have IRESes)
two general types of multiple AUGs:
if Kozak sequence is very weak
AUG/UGA combo
Weak Kozak Sequence
ribosomal subunit scanning for AUG is then called ‘leaky scanning’ → results in upstream ORF
ribosomal subunit moves to the second or third AUG (usually in frame)
resulting proteins have different N-terminus which is the part that usually encodes a signal sequence (responsible for cell destination)
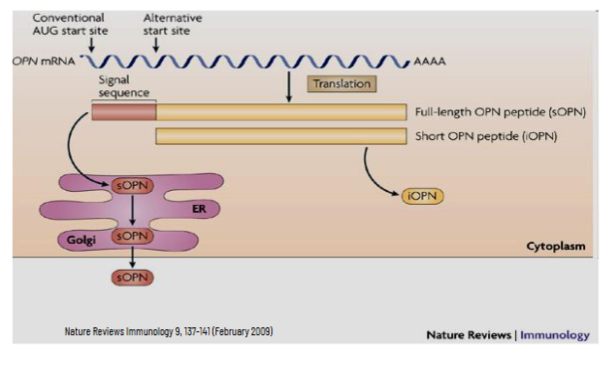
AUG/UGA Combo
short open reading frame between 5’ end of mRNA and beginning of the main coding sequence - upstream open reading frame (uORF)
coded polypeptide is usually not functional
this stalls/decreases the translation of the actual downstream gene by “trapping” that scanning ribosome and causing it to drop down from the mRNA, before it reaches the AUG of the main protein coding sequence
basically, it makes a small peptide to stall the translation of the protein of interest since it takes energy and resources
GCN4
AA starvation
GCN4
type of AUG/UGA combo, is yeast amino acid synthesis genes
normal growth: lots of 43S preinitiation complexes; translation from uORGs
the ribosomes that terminate translation on uORFs cannot resume scanning
reinititation is very low from ‘real’ AUG5
low level translation of GCN4
AA Starvation
type of AUG/UGA combo where fewer 43S preinitiation complexes formed by phosphorylation of eIF2
more initiation from the ‘real’ AUG5
more GCN4 is produced (translation factor)
high levels of translation of amino acid biosynthetic genes
Features of 5’ UTR
they determine the impact of an uORF on the translation of the main ORF
sequence context makes it possible for the formation of RNA secondary structures
the length of the sequence upstream, and the sequence context of the uAUG both influence the ‘visibility’ of the start codon (hides the right AUG)
length of uORF and the sequence context of the uORF stop codon influence the competence of ribosomes to reinitiate on the main ORF once they have terminated translation (affects efficiency of assembly at main ORF)
uORF-encoded peptide can cause ligand-dependent stalling of the ribosome
sequences downstream of the uORF stop codon can influence both reinitiation and the stability of the mRNA
Internal Ribosome Entry Sites (IRES)
in the 5’ UTR
enables cap-dependent translation - bypass requirement for binding of eIF4E (binds methyl cap) and eIF4G (binds PABP) factors
is around 450 nucleotides long with complex secondary structure; rich in Us and Cs
43S preinitiation complex binds to IRES upstream from the ‘normal’ AUG and scans downstream from there
IRES-specific binding proteins are required
locates AUG in Kozak for initiation
Why is IRES important?
viral infections: cap-dependent translation can be inhibited in cells due to cleavage of eIF4G (an adaptor to bridge the 40S subunit) by viral protease → outcome is only viral mRNA is translated
mitosis: decreases cap-dependent translation
cell in stress conditions: inhibits cap-dependent translation since translation factors get phosphorylated and turned off
some cellular mRNAS have IRES AND cap
coding for some of stress induced proteins, some homeodomains proteins, some growth factors and apoptosis-associated proteins
their translation is not inhibited under conditions when overall (cap-dependent) protein synthesis is temporarily shut down
selective activation of IRES mediated translation
basically controls gene expression through cap-dependent translation, regulation of balance between apoptosis and cell division by translating a different sets of proteins
Review of Protein Coding Gene, mRNA, and ORF
this makes coding sequence
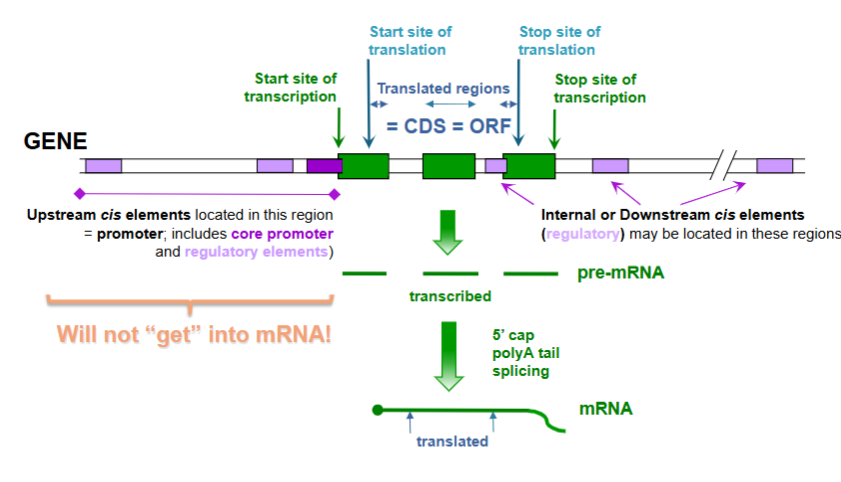
Generic Structure of Eukaryotic mRNA
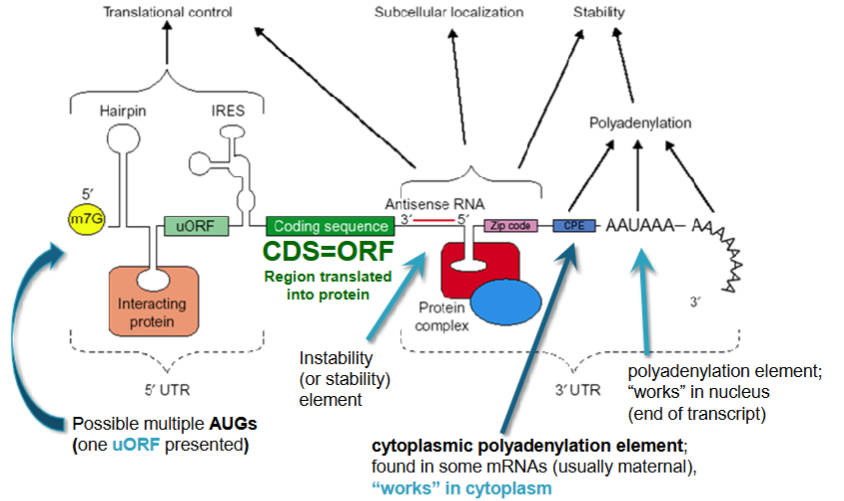
Cytoplasmic Polyadenylation Element (CPE)
found in the 3’ UTR of some mRNA (usually maternal), works in the cytoplasm
is the signal for the length of the poly-A tail to be longer or shorter
different from poly-adenylation signal (PAS) which affects cleavage and poly-adenylation in the nucleus
Prokaryotic Elongation
after intitiation, tRNAi Met is on P site
amino-acyl-tRNA couples with elongation factor thermo-unstable (EF-Tu) and GTP into ternary complex, binds to A site on ribosome and pairs with codon on mRNA. this is catalyzed by GTP
peptide bond forms between previous amino acid and new one by peptidyl transferase reaction-previous amino acid is transferred to the amino acid carried by amino-acyl-tRNA which now becomes peptidyl-tRNA still at A site; unloaded tRNA is still at the P site
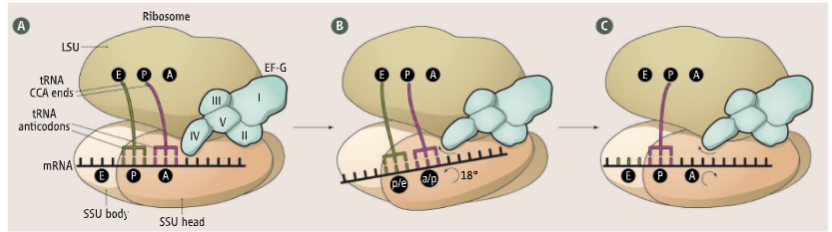
ribosome moves 3’ to move another codon into the A site (translocation is catalyzed by EF G (Pro) using GTP energy
peptidyl-tRNA shifts to P site, empty tRNA is now at E site, A site is empty so new amino acid can come in
new amino-acyl-tRNA with GTP, EF-Tu, binds to A site in a GTP-dependent manner
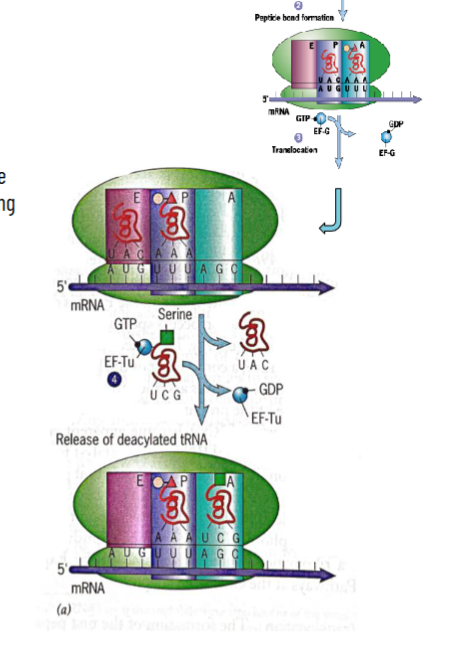
EF-G During Elongation
small subunit head is twisted to facilitate codon translocation
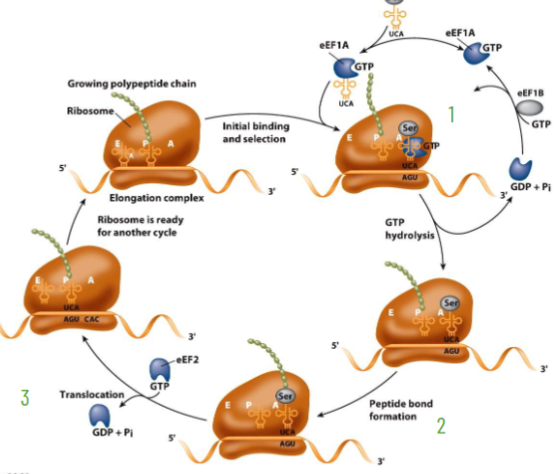
Eukaryotic Elongation
amino-acyl-tRNA, eEF1A-eEF1B and GTP binds to A site
peptide bond forms peptidyl-tRNA, still at the A site, unloaded tRNA still at the P site
ribosome translocates 3’ catalyzed by eEF2 (Eu) using GTP energy
Puromycin
causes premature translation termination by acting as an aminoacyl-tRNA analog that is incorporated into the growing polypeptide chain - no carboxyl group available for peptide bond to form with amide from previous amino acid
Peptidyl Transferase
catalyzes the transfer of the growing peptide chain to the incoming activated amino acid and makes the peptide bond
tRNA makes direct contact with both 16S and 23S rRNAs in ribosome, and rRNAs are highly conserved way more than ribosomal proteins
ribosomal peptidyltransferase center is for: peptide bond formation and nascent peptide release during elongation and termination
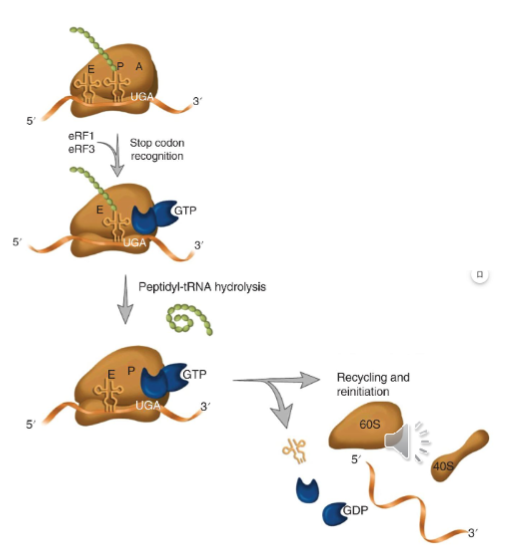
Termination
there is no tRNA at stop codons (UGA, UAA, UAG)
the stop codon in the A site is recognized by the release factors
with no charged tRNA, nascent peptide gets released once the ester bond linking the polypeptide to the P-site tRNA is hydrolyzed
ribosome can be disassociated and recycled