Enzyme Regulation and Allostery (Ch. 15)
1/33
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
34 Terms
The velocity of a reaction is typically controlled by?
the concentrations of substratesand cofactors (cofactors: metal ions or organic coenzymes that participate in some enzyme reactions)
As the product of the reaction accumulates...
-apparent rate of product formation will slow
-due to increasing rate of reverse reaction
-dirrectly dependent on [P]
What determines the amount of enzyme present at any moment?
-genetic regulation of enzyme synthesis and decay
Enzyme regulation through reversible, noncovalent binding of metabolic effectors at sites other than the active site is called?
-allosteric regulation
Enzyme activity can also be regulated through covalent modification
-reversible attatchment to amino acid side chains of enzyme
Examples of covalent modification?
Phosphorylation
also, the enzymes that introduce these modifications can themselves be regulated
Enzyme activity can also be regulated through zymogens (proenzymes)
-these are inactive precursors from which active enzymes can be generated by proteolytic cleavage
-this is irreversible
Examples of zymogens?
-insulin is made as an inactive 86 amino acid precursor (proinsulin)
-removal of residues 31-65 generates active form which has two chains (A- and B- chain) and three disulfide bonds
-the cleaved connecting peptide is called C-peptide
-also the zymogen of chymotrypsin is chymotrypsinogen, which is activated first by trypsin
Enzyme activity can also be regulated through isozymes
-enzymes with slightly different subunits
-subunits are homologous
-different isozyme subunits have different kinetic properties
Example of isozymes?
-five isomers of lactate dehydrogenase
-A4 to A3B .... AB3 to B4
Allosteric enzymes catalyze committed steps, many allosteric enzymes are susceptible to?
-feedback inhibition by the end product of the metabolic pathway
Do allosterically regulated enzymes follow Michaelis-Menten model for enzyme kinetics?
no
Substrate concentration curves for allosteric enzymes are?
sigmoid (S-shaped)
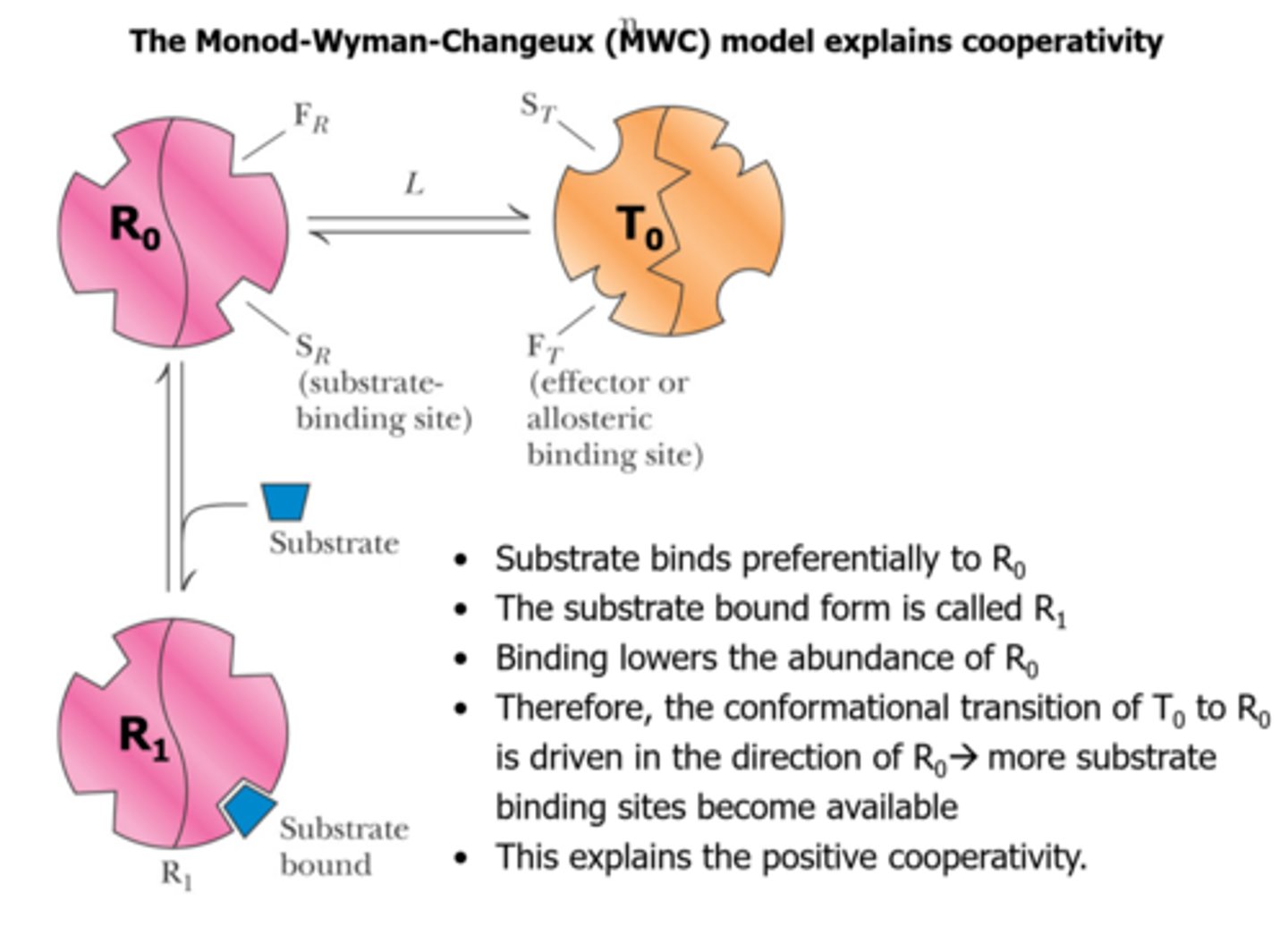
Substrate binding is said to be cooperative, meaning?
binding of one molecule of substrate facilitates the binding of subsequent molecules
Regulatory enzymes are oligomeric
-each subunit in an allosteric enzyme has a substrate-binding site and an effector-binding site
Allosteric effector (inhibitor or activator) usually has ________________ structural similarity to the substrate
little or no
Regulatory effects depend on ________________________ occurring in the enzyme as a result of effector binding
conformational changes
Conformational changes in allosteric enzymes are the basis of changing... ?
affinity for the various ligands
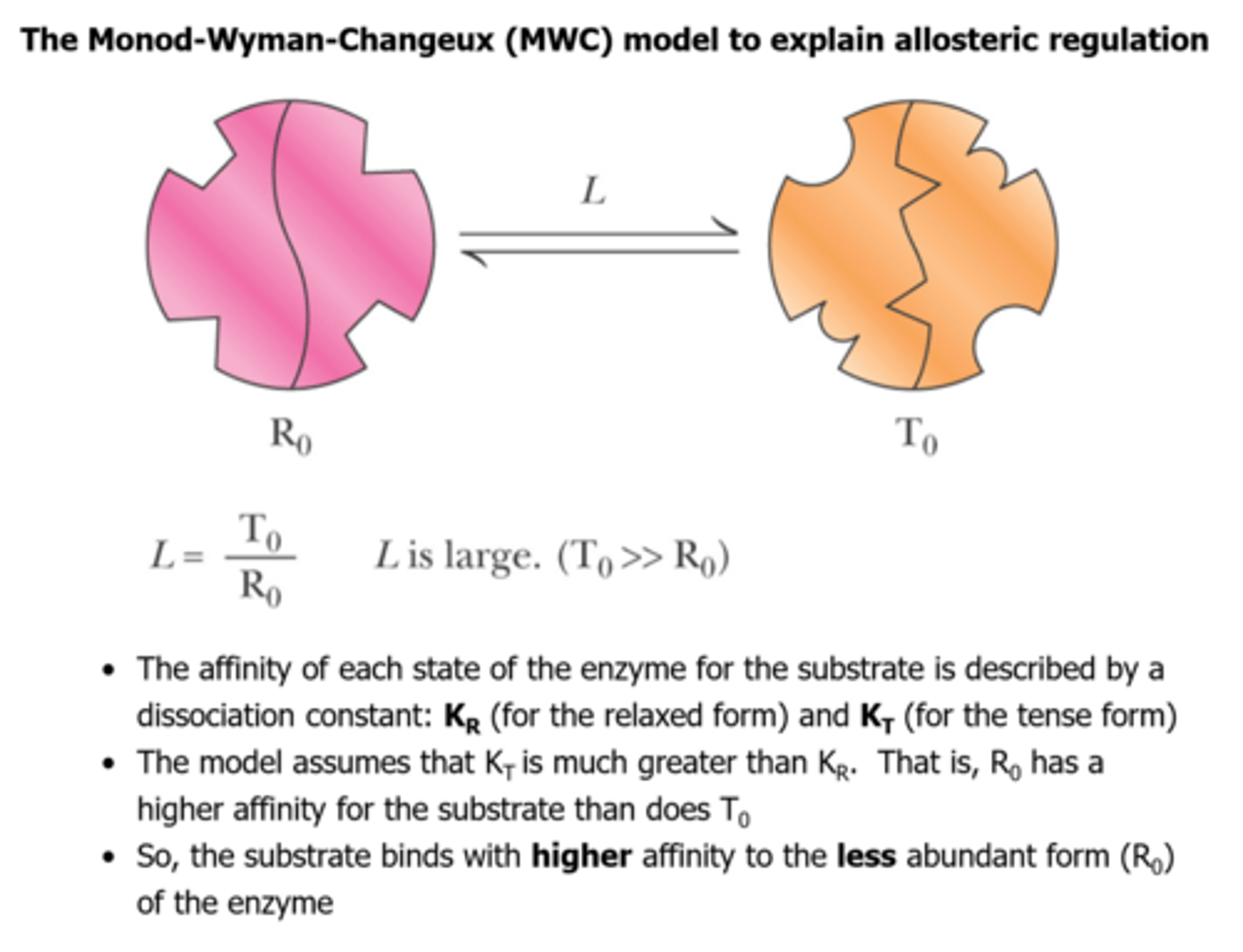
Monod-Wyman-Changeux model (MWC) postulates that:
-allosteric enzymes are oligomeric
-allosteric enzymes can exist in (at least) two conformational states, called R and T
-different conformations have different affinities for the various ligands
-'L' is the equillibrium constant for the R - T equillibrium defined as L = T0/R0, if L is large, and S binds only to R, substrate binding is cooperative
-concentration of ligand giving half-maximal saturation is defined as K0.5
The Monod-Wyman-Changeux model (MWC) is based on linked equllibria between.... ?
conformational state and ligand-binding properties of allosteric proteins (concerted model)
The Monod-Wyman-Changeux (MWC) model to explain allosteric regulation
Oligomeric enzyme has two conformational states:
R (relaxed)
T (tense, or taut)
-In each protein molecule, all subunits have the same conformation
-In the absence of ligand, the two states are called R0 and T0
-The equilibrium constant (L) for the T0/R0 equilibrium is large, that is T0 predominates
The Monod-Wyman-Changeux (MWC) model to explain allosteric regulation CONTINUED
The MWC model cannot explain?
-negative cooperativity in substrate binding
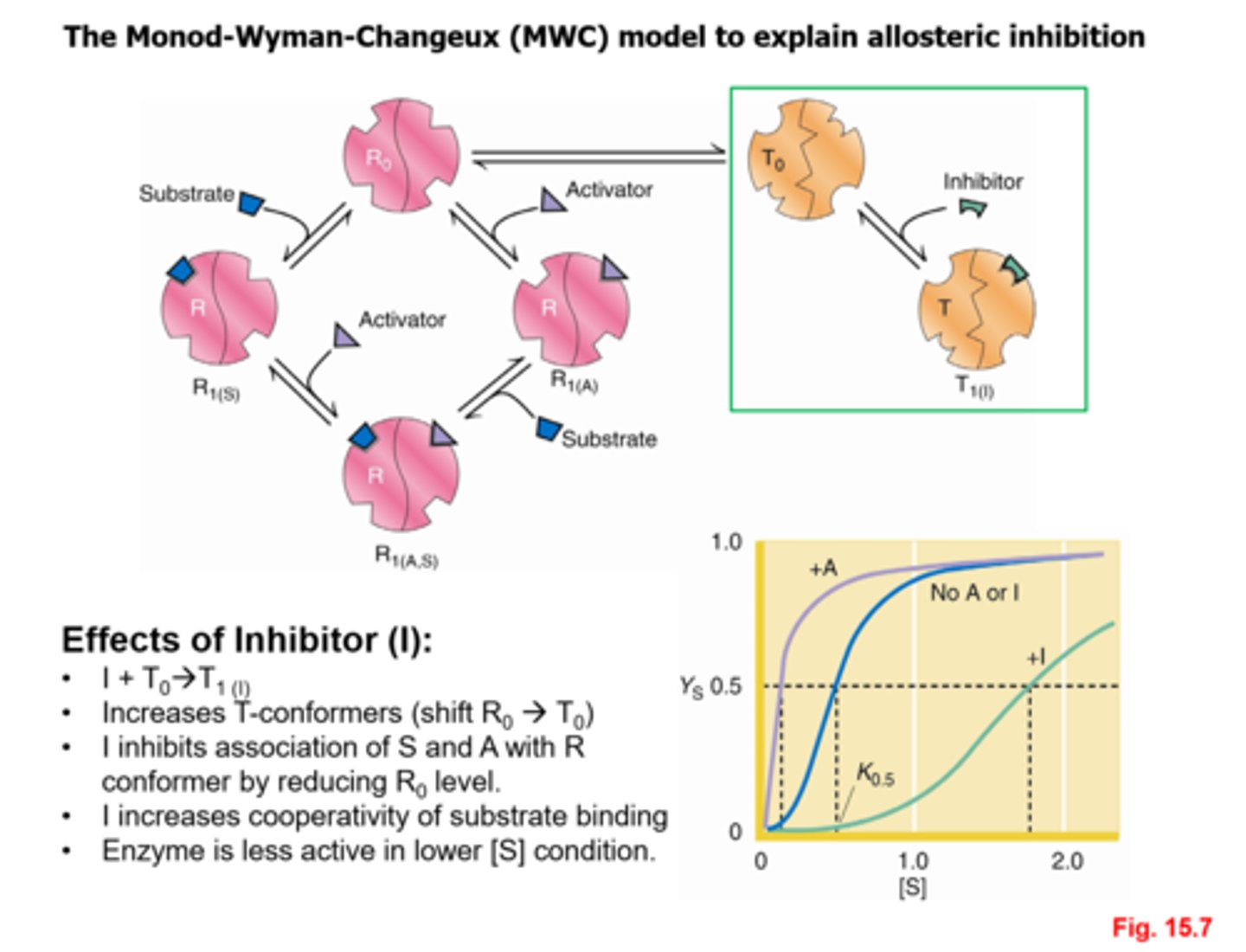
How does the MWC model explain allosteric activation?
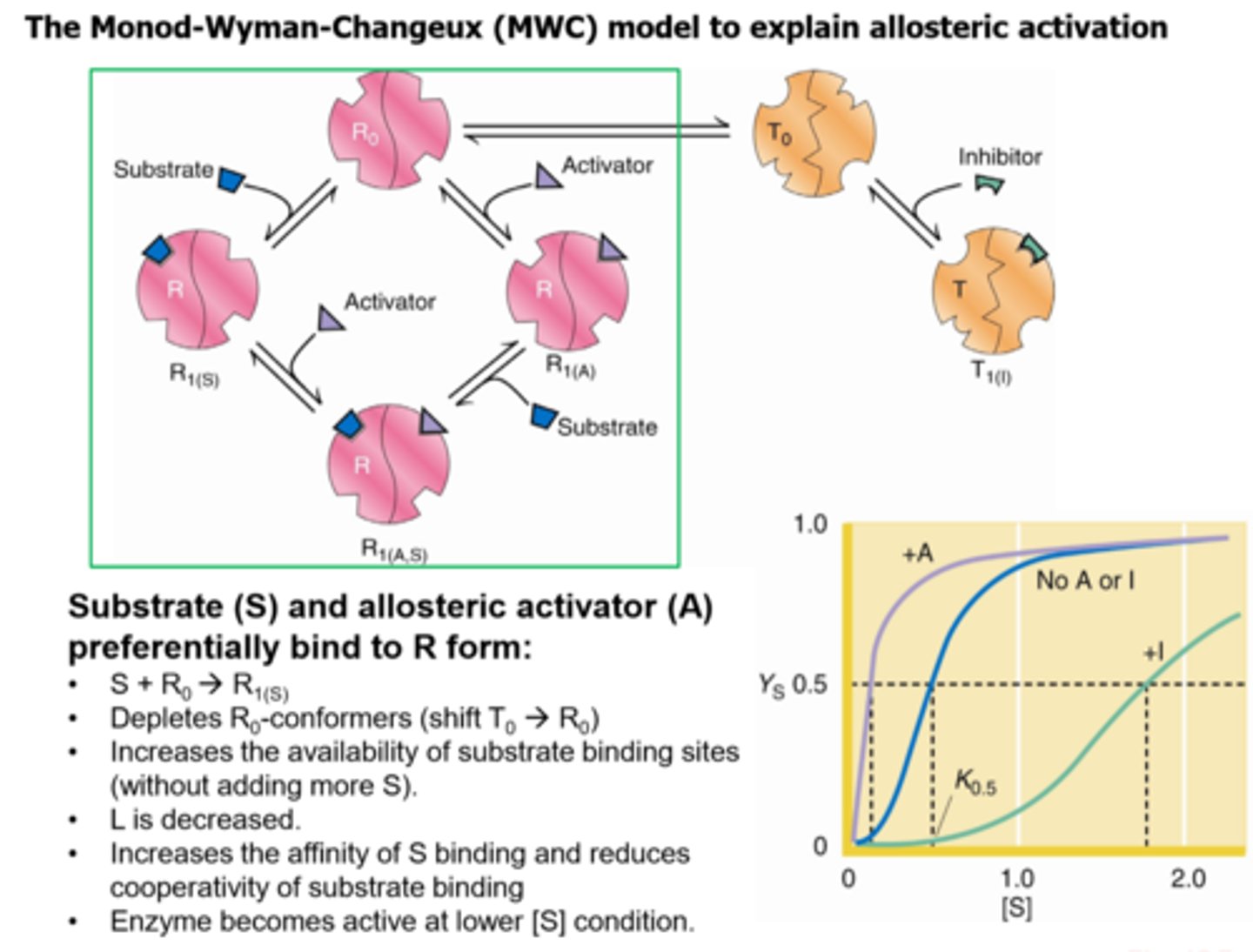
How does the MWC model explain allosteric inhibition?
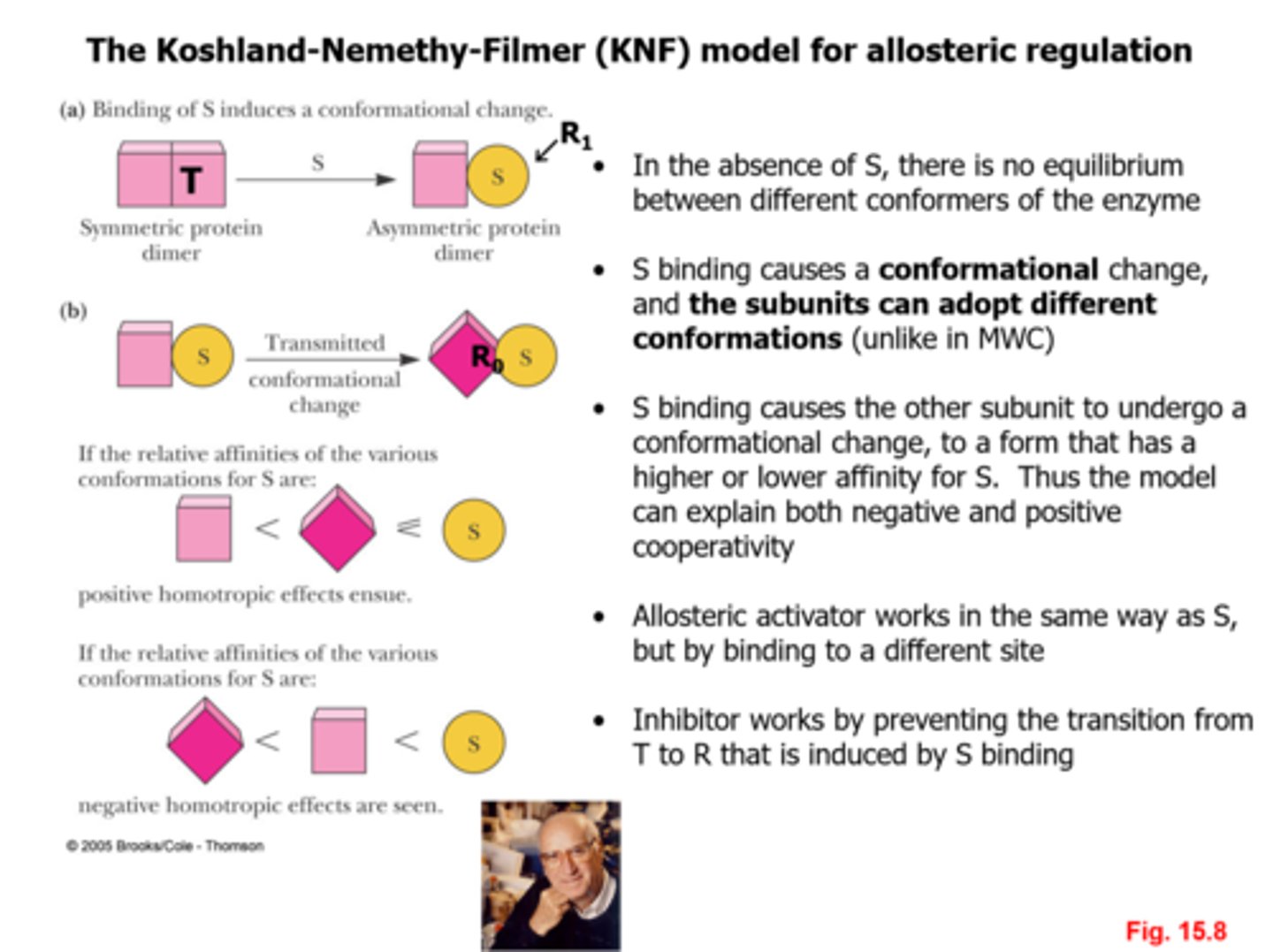
The Koshland-Nemethy-Filmer (KNF) model for allosteric regulation is based on?
-ligand-induced conformational (sequential) changes in allosteric proteins
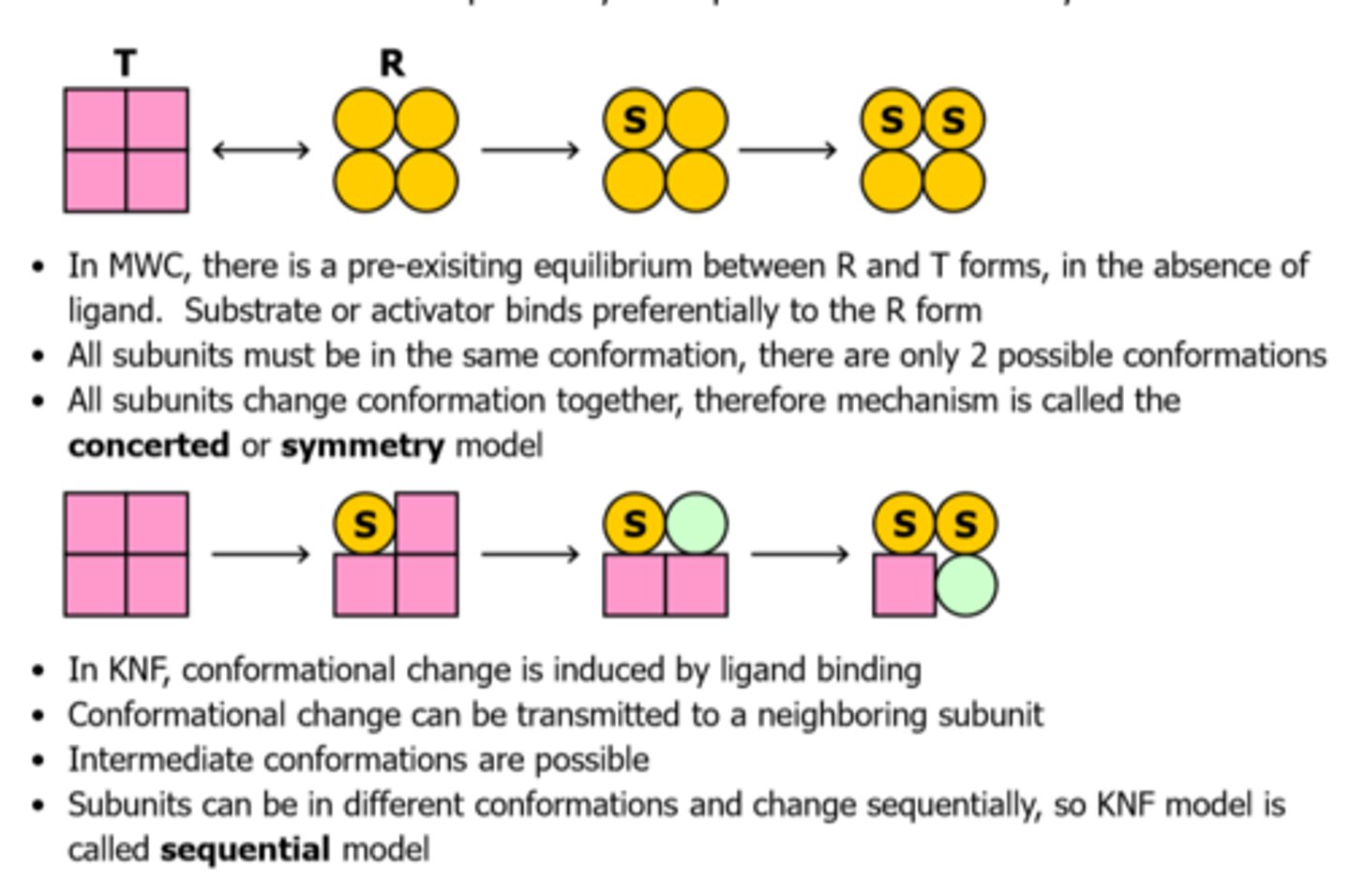
Key differences between the MWC and KNF models?
In MWC:
-there is pre-existing equillibrium between R and T forms
-substrate or activator binds preferentially to R form
-all subunits must be in the same conformation, there are only 2 possible conformations
-all subunits change conformation together, therefore mechanism is called the concerted or symmetry model
In KNF:
-conformational change induced by ligand binding
-change transmitted to a neighboring subunit
-intermediate conformations are possible
-called sequential model
MWC and KNF models comparison picture
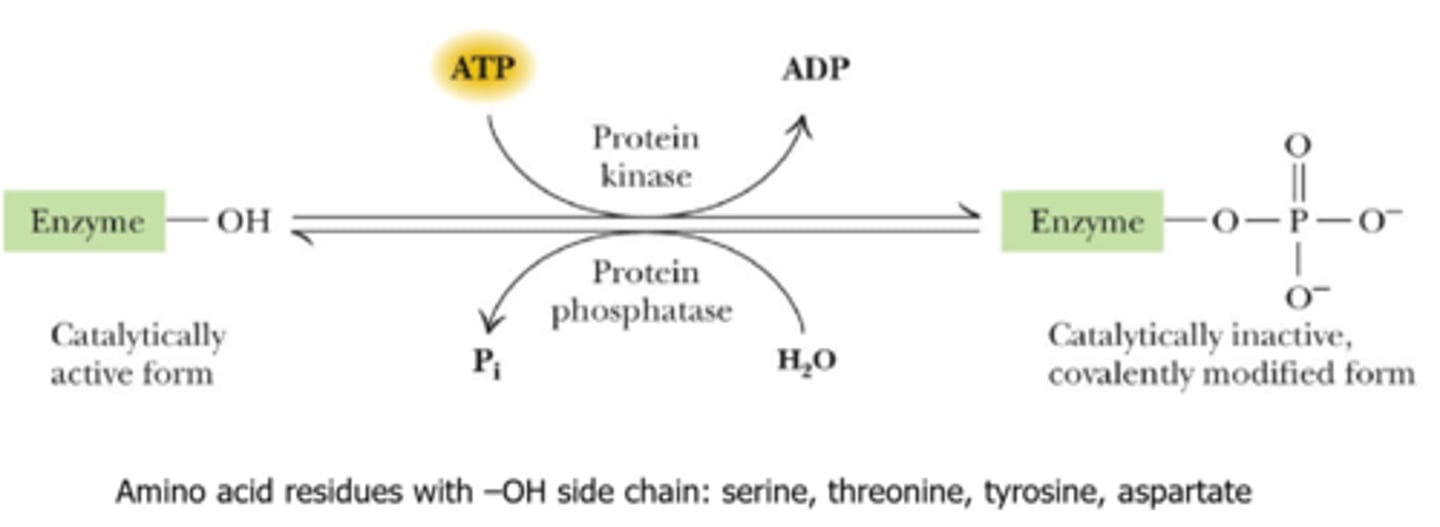
Protein kinases do what?
catalyze ATP dependent phosphorylation of target protein
The prominent means of metabolic regulation?
covalent modification through reversible phosphorylation
Target amino acid residues for protein kinase phosphorylation?
amino acids with -OH side chain:
-serine
-threonine
-tyrosine
-aspartate
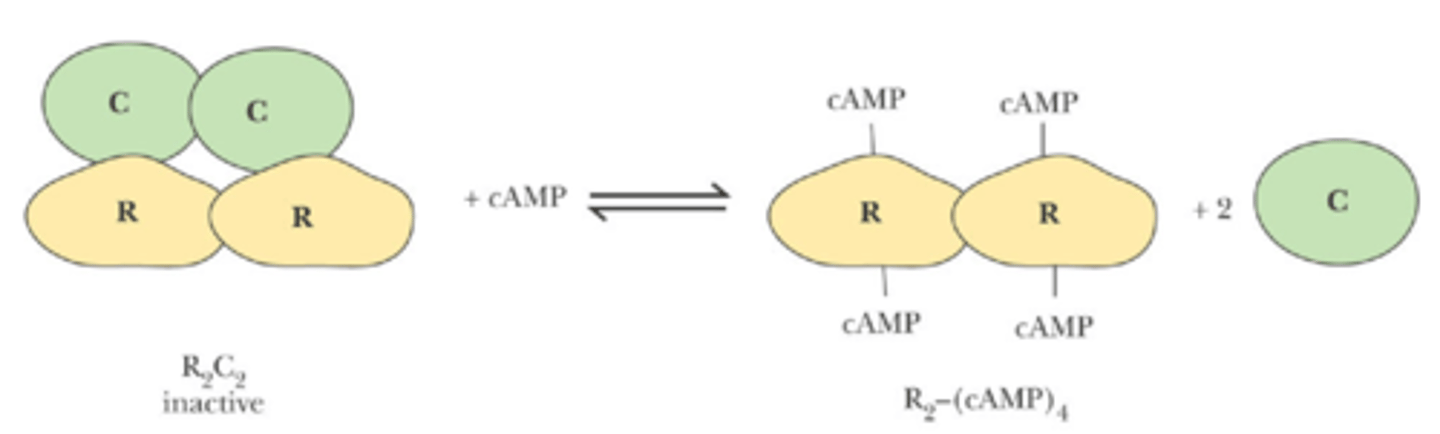
Other form of covalent modification that regulate protein function?
-catalytic (C) subunit of cAMP-dependent protein kinase (PKA) is kept inactive by the regulatory R subunit, binding of cAMP to the R subunit releases the active enzyme
Glycogen phosphorylase?
a paradigm of allosteric regulation and covalent modifiaction through reversible phosphorylation
Important theme in metabolic regulation:
Covalent modification _____________ allosteric regulation.
overrides