Exam 2 - Lectures 7+8
1/108
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
109 Terms
mRNA translation
process where mRNA is decoded to produce proteins
Major features of mRNA translation
occurs in all parts of the cell cycle
produces functional peptides and proteins
most of other RNA types are not translated
in prokaryotes, occurs in the cytosol
in eukaryotes, occurs in the cytosol and on ER membrane
codons
triplets of nucleotides that code for a specific amino acid - basis of genetic code
How many standard codons are there?
64 - 4³
61 code for amino acids
3 code for stop signals
Anticodons
nucleotide sequences that are complementary to their corresponding mRNA codon sequence
tRNA
adaptor molecules that contain the anticodon and link anticodon to codon to
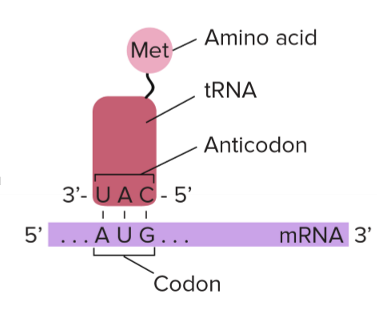
what direction does mRNA translation happen?
5’ to 3’
codon table
translates the genetic code into amino acids
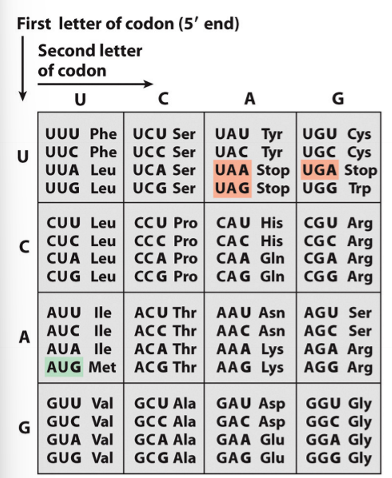
the genetic code is degenerate which means
a single amino acid may be coded for by more than one codon (except Met and Trp)
The genetic code is nearly universal which means that
it is used by prokaryotes and eukaryotes across species
codon usage bias
codon frequencies and anticodon/tRNA frequencies vary between organisms
what causes codon usage bias?
mutations
each tRNA molecules carries an ____ and recognizes ___
carries an activated amino acid and recognizes a specific mRNA codon
How are 32 tRNA sufficient for 61 codons?
Because of wobble base pairing between tRNA anticodon and mRNA codon
where does wobble base pairing in tRNA anticodon with mRNA codon occur?
third nucleobase
what do most codes start with?
AUG - methanine
reading frame
way of dividing the sequence of nucleotides into triplets, is established by the first codon read in the sequence
if the first nucleotide is skipped, what happens?
the reading frame is altered
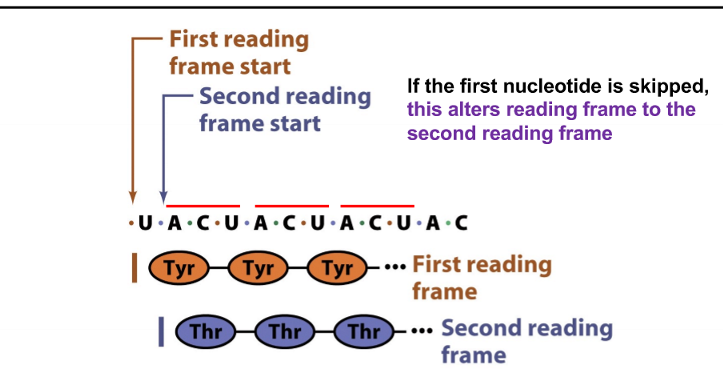
if there are three nucleotides skipped, what happens to the reading frame?
nothing, since amino acids are produced by nucleobase triplets
frameshift mutations
change the frame - insertions or deletions of a number of nucleotides that is not divisible by 3
slippery mRNA sequences
small stretches of codons that cause translational frame shifts (usually contains a lot of U and A nucleobases)
What causes slippery mRNA sequences?
tRNAs slipping (incorrect base pairing)
degenerate code
more than one codon codes for one amino acid - allows most minor mutations to still code for the same amino acid
NOT ALL MUTATIONS INFLUENCE PROTEINS MADE
silent mutations
from degenerate code, differ in the DNA nucleotide sequence but code for the same protein
nonsense mutations
adds stop codon early
missense mutations
change in DNA nucleotide sequence that changes the protein made
ribosomes
perform mRNA translation (protein synthesis)
five steps of mRNA translation
tRNAs are loaded/charged with an activated amino acid
tRNA is loaded with an amino acid (aka it is aminoacylated)
Translation initiation
mRNA and aminoacylate tRNA bind to the ribosome
Translation elongation
cycles of aminoacyl-tRNA binding and peptide bond formation occur until a stop codon is reached
Translation termination
mRNA and protein dissociate, ribosome recycled
Protein folding and posttranslational modifications
catalyzed by a variety of chaperones and enzymes
key players in bacterial mRNA translation
transfer RNA (tRNA)
amino acids
aminoacyl tRNA synthetase
Messenger RNA (mRNA)
30S ribosome
50S ribosome
Energy (ATP, GTP)
Initiation factors
elongation/T factors
Release factors
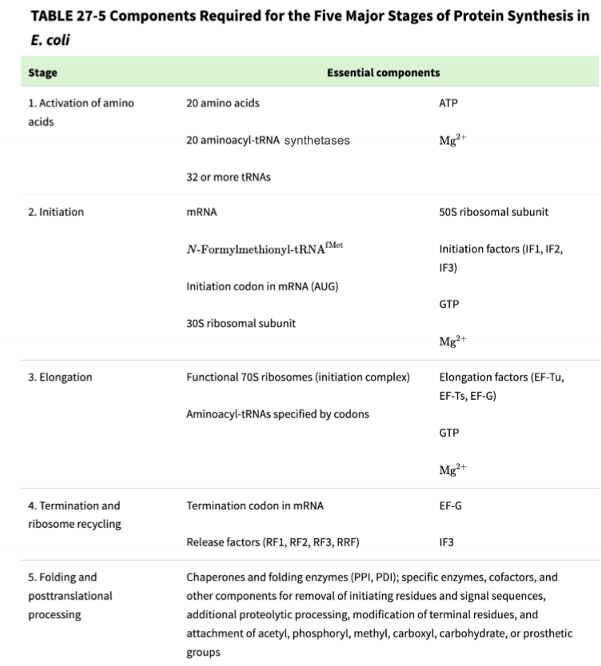
stage 1: charging tRNA with an amino acid
part a - forming aminoacyl adenylate
part b - loading the activated amino acid onto the tRNA to make a charged tRNA (called an aminoacyl-tRNA)
aminoacyl tRNA synthetases (aaRS)
enzymes that help activate amino acids and them load them onto tRNA, there are 20 different aaRS to make 32 different types of aa-tRNAs
aaRS interacts both at the amino acid arm and the anticodon region of the tRNA to provide specificity
the aaRS catalytic domain associates with the correct amino acid
the aaRS anticodon recognition domain associates with the correct tRNA anticodon sequence
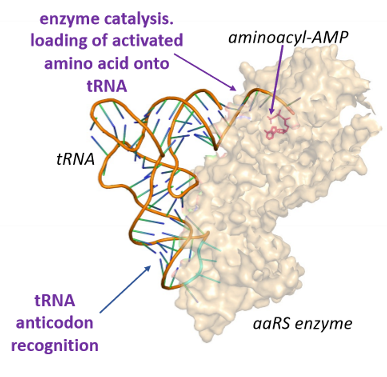
ribosome
machinery that carries out mRNA translation
catalyzes protein synthesis using peptidyl transferase enzyme activity
composed of a large subunit and a small subunit
in bacteria the subunits are 50S and 30S
in eukaryotes the subunits are 60S and 40S
ribosome is a mixture of many different proteins as well as different rRNA molecules
Protein subunits are stabilized by rRNA and vice-versa
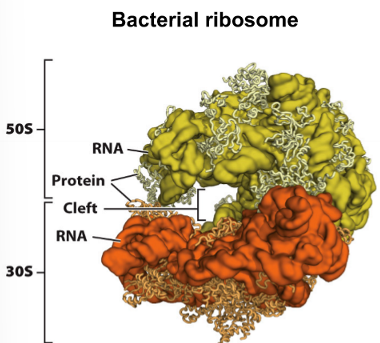
3 sites of a ribosome
A site - aminoacyl
P site - peptidyl
E site - exit
A site of ribosome
site for incoming aminoacyl tRNA
P site of ribosome
holds the tRNA with the peptide attached, which is to be transferred to the new amino acid residue in the course of the peptidyl transferase reaction
E site of ribosome
the third and final binding site for the tRNA in the ribosome during translation
where does initiation initiating tRNA (fMet-tRNA) enter?
P site
where does all of the tRNAs after the initiating tRNA enter?
A site
how does the ribosome know where to bind to mRNA?
specific sequences between the promoter and the gene
Shine-Dalgarno sequence in bacteria
5’ cap and Kozak sequence in eukaryotes
stage 2: translation initiation
a. the small ribosome subunit binds to multiple initiation factors, which help recruit mRNA
b. 16S rRNA binds to the ribosome and the Shine-Dalgarno sequence of mRNA to stabilize the mRNA/30S ribosome structure
c. IF2-GTP recruits tRNA (fMET) and associates with 30S ribosome/mRNA; tRNA (fMet) binds to P site
d. the large ribosome subunit associates with the IF/mRNA/ribosome/tRNA complex and releases IFs to form the full ribosome/mRNA/tRNA complex
initiation factors (IF)
proteins that bind to the small ribosome subunit during translation initiation
Stage 3: translation elongation
composed of 3 steps that all happen in the ribosome
a. decoding
b. peptide bond formation
c. translocation
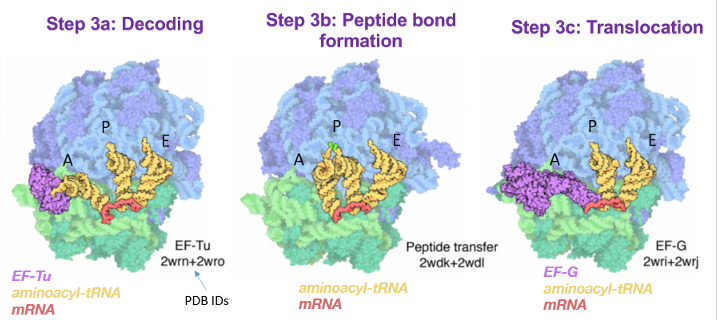
Step 3a - decoding
another charged aminoacyl-tRNA bound to GTP and elongation or T factors (Tu) enters the A site of the ribosome
binding and peptide bond formation are accompanied by GTP→ GDP hydrolysis and release of the Tu-GDP complex from the ribosome
Tu-GTP complex is regenerated in a process requiring other T factors and GTP
Results in a change in the conformation of the 2nd amino-acryl tRNA that pulls its aminoacyl end into the P site
step 3b - peptide bond formation
a peptide bond is formed between the alpha nitrogen of one amino acid and the carbonyl carbon of another amino acid
peptidyl transferase activity that catalyzes peptide bond formation resides in the 23D rRNA (a ribozyme) rather than in any protein compartments of ribosomes
driven by favorable entropy change
the reaction does NOT involve chemical catalysis but may be modulated by conformation changes in the active site what can be induced by protonation
step 3c - translocation
the ribosome shifts the next codon towards the 5’ end of the mRNA
requires EF-G (translocase) and the energy from GTP hydrolysis
EF-G binds the A site and displaces the peptidyl-tRNA
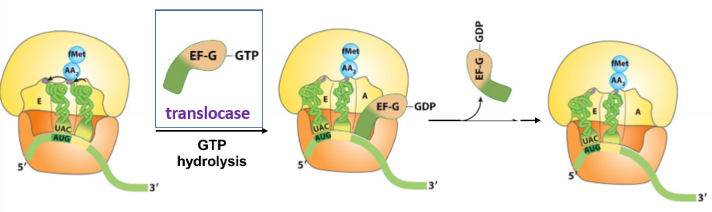
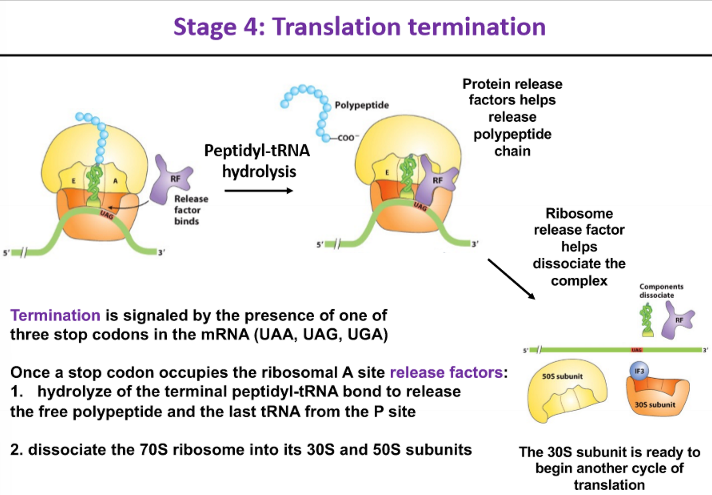
stage 4 - translation termination
signaled by the presence of one of the 3 stop codons in the mRNA
once a stop codon occupies the ribosomal A site, release factors:
hydrolyze the terminal peptidyl-tRNA bond to release the free polypepride and the last tRNA from the P site
dissociate the 70S ribosome into its 30S and 50S subunits
polysomes
ensembles of 2+ consecutive ribosomes that translate mRNA into proteins
T/F - DNA transcription and mRNA translation can occur in cells as coupled processes
TRUE
Stage 5 - protein folding and posttranslational modifications
polypeptides leave the ribosome through an exit tunnel
the polypeptide chain is folded and processed into its biologically active form
proteins can fold spontaneously or have chaperonins assist
post translational modification
covalent modification of amino acids
mRNA translation is the most energy-consuming process in the cell
must be tightly controlled by ATP/GTP availability in order to maintain homeostasis
How many AP is required to make a __ long peptide?
= (# of peptides x 4)
2 ATP per aa for step 1
1 GTP for initiation
(# of aa - 1) for elongation
1 GTP for termination
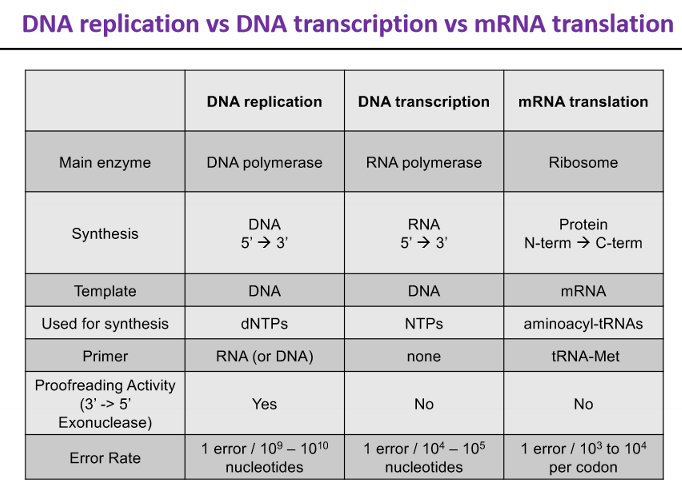
DNA replication vs DNA transcription vs mRNA translation
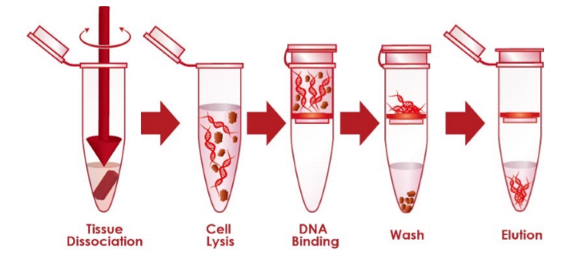
nucleic acid (DNA or RNA) extraction
method to extract and purify nucleic acids - separating nucleic acids from other cellular components
5 basic steps of nucleic acid extraction
collect and resuspend the sample
disrupt the cellular structure to create a lysate
bind the nucleic acids to a purification matrix
wash out other cellular components from the matrix
elute the nucleic acid from the purification matrix
genomic DNA
total genetic info of an organism
plasmid DNA
small circular DNA usually external from genomic DNA
synthetic DNA
artificial/synthesized DNA
agarose gel electrophresis
how to visualize nucleic acids
nucleic acids separated based on size and length
DNA and RNA are negatively charged so they move towards the positively charged anode
technique to separate nucleic acids
nucleic acids separated by applying an electric field to move the charged molecules through an agarose gel matrix
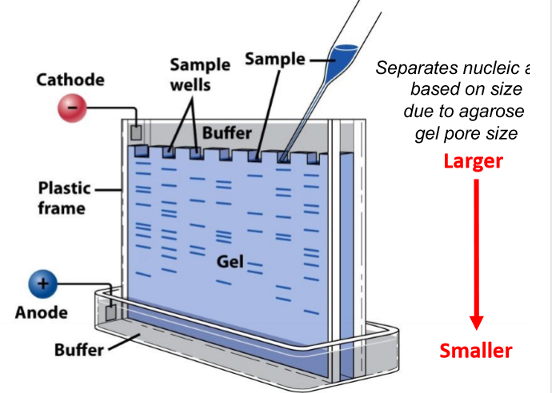
higher percentage agarose gels are better are separating __ nucleic acid fragments while lower agarose gels are better at separating ___ nucleic acid fragments
high % = smaller fragments, lower % = larger fragments
intercalating agents - ethidium bromide
way to visualize nucleic acids
becomes fluorescent upon binding to double stranded DNA or helical RNA
not specific
hybridization probes (complementary fragments of DNA or RNA)
way to visualize nucleic acids
the probes are fluorescently labeled and bound to target DNA or RNA (binding specificity)
better than ethidium bromide because this is
able to be used on RNA - while ethidium bromide binds to double stranded DNA/helical RNA, most RNA is not double helix
has sequence specificity
blotting
technique where biomolecules are resolved in a gel matrix, transferred to a solid support, and detected with a specific probe
Northern blot
RNA
Southern Blot
uses electrophoresis to separate DNA by size and detect it with a hybridization probe complementary to the target sequence
what technique should you use if you don’t have a lot of nucleic acid sample to test?
PCR
DNA amplification
process where DNA is enzymatically copied to generate millions of identical copies of the parent DNA molecule
reasons why you might want to amplify DNA (same reasons for nucleic acid extraction)
forensic analysis
genome sequencing
paternity/maternity/ancestry testing
medical testing
pathogen testing
PCR - polymerase chain reaction
in vitro DNA amplification method that takes advantage of mechanisms behind cellular DNA replication
ingredients for standard PCR reaction
template (genomic, plasmid, or synthetic DNA)
primers (usually DNA fragments since they are more stable)
Thermostable DNA polymerase
Buffer (has ions needed for enzyme)
dNTPS (the four types of DNA nucleotides)
oligoucleotides
short, single-or double stranded DNA or RNA molecules
primer
short nucleic acid sequence that provides a starting point for DNA synthesis
For PCR, the terms primer and oligo are often used ___
interchangeably
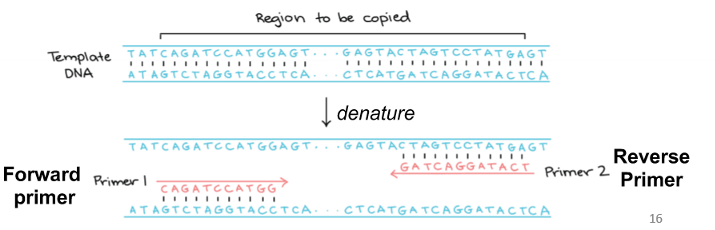
forward primer
attaches to anti-sense strand
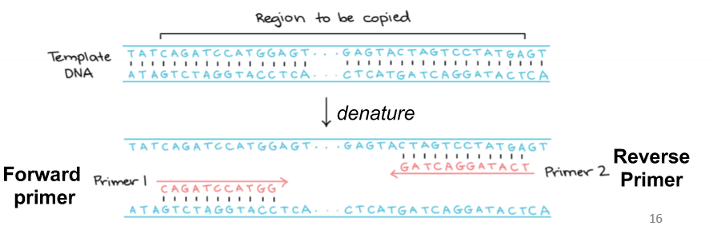
reverse primer
attaches to sense strand
Taq/Pfu DNA polymerase
2 different thermostable enzymes commonly used in PCR
enables running the PCR at high temperature to completely denature DNA
PCR steps
denature the template DNA helix by heating the mixture over 95 C
anneal the DNA primer to the template by cooling to 50-70 C
polymerize the dNTPs to the primer using thermostable DNA polymerase by heating to 72 C (5→3 elongation by synthesis of new DNA strands)
allow for exponential DNA amplification by repeating the denature and anneal cycles to double the number of copies in each cycle
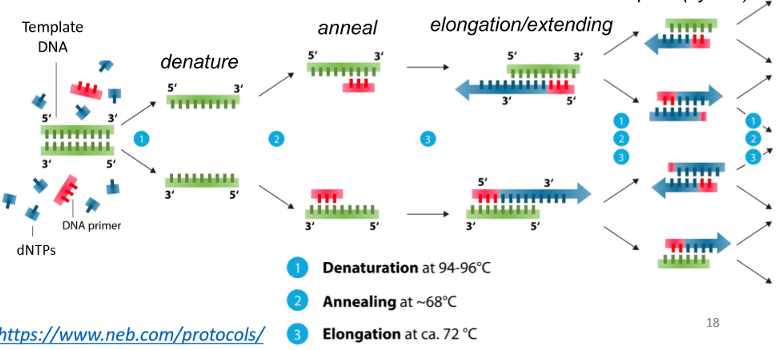
the number of DNA molecules produced is doubled in each PCR cycle
so after n cycles, you have 2^n copies of DNA
DNA replication vs PCR
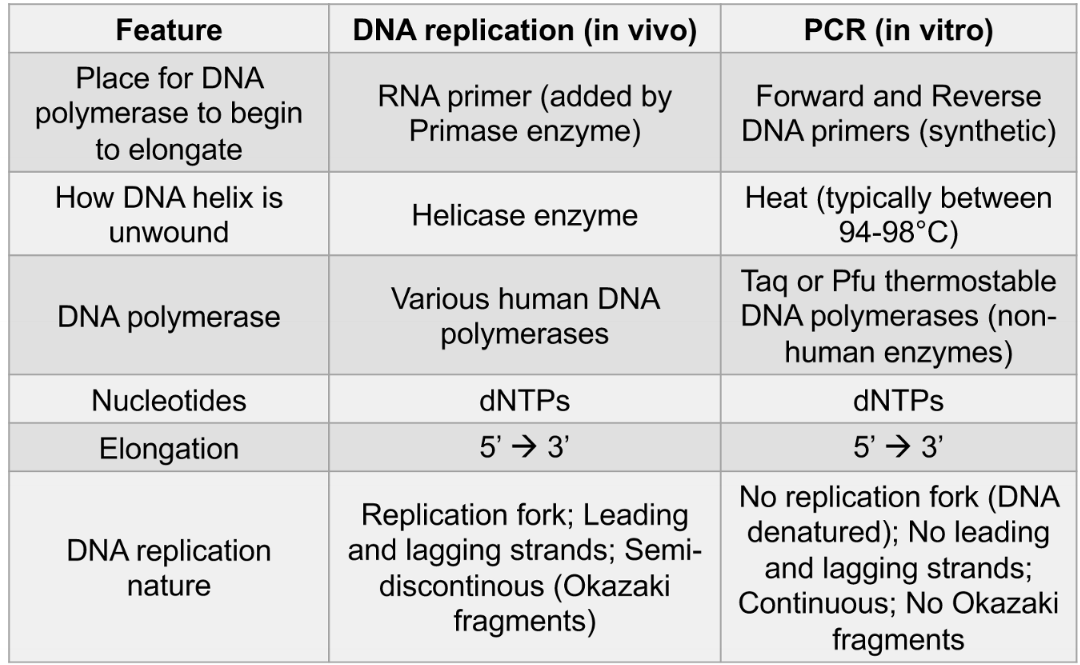
What technique should you use if you have RNA for a template?
RT-PCR (reverse transcription)
RT-PCR
technique enabling reverse transcription of RNA to DNA using modified PCR
Ingredients for standard RT-PCR
RNA template
Primers (usually DNA)
Thermostable RNA dependent DNA polymerase (reverse transcriptase)
thermostable DNA dependent DNA polymerase (Taq or Pfu)
dNTPs
reverse transcriptase
enzyme used to generate complementary DNA (cDNA) from an RNA template (process is called reverse transcription)
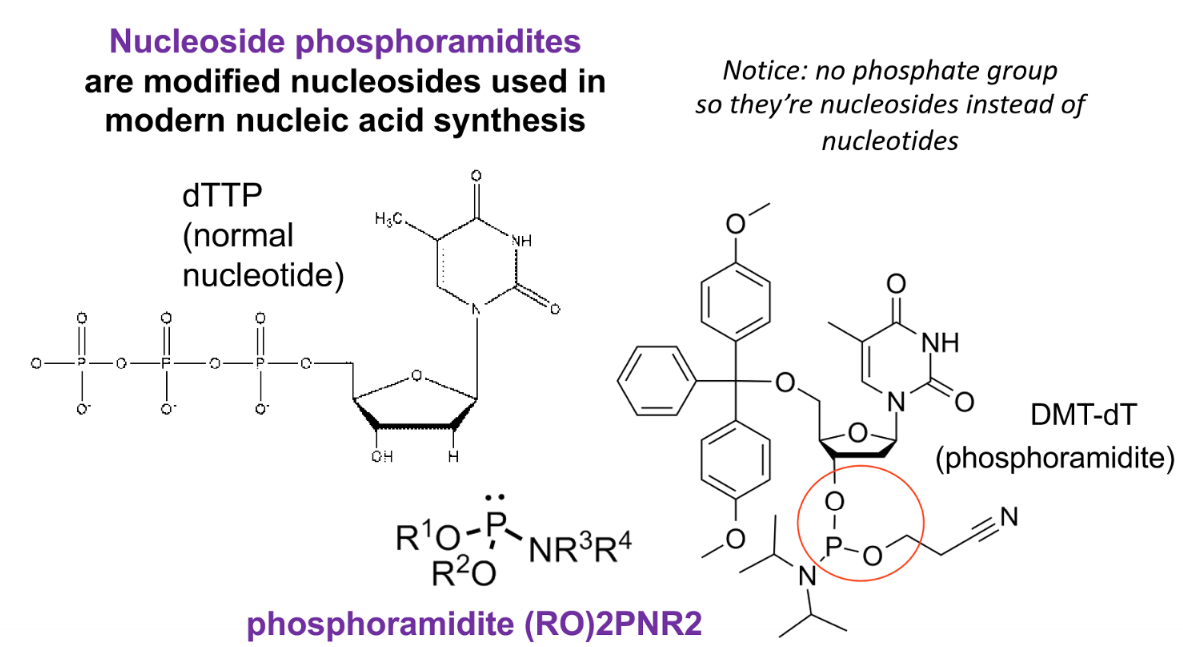
phosphonamidite method
method of chemical synthesis
nucleoside phosphoamidites
modified nucleosides used in modern nucleic acid synthesis
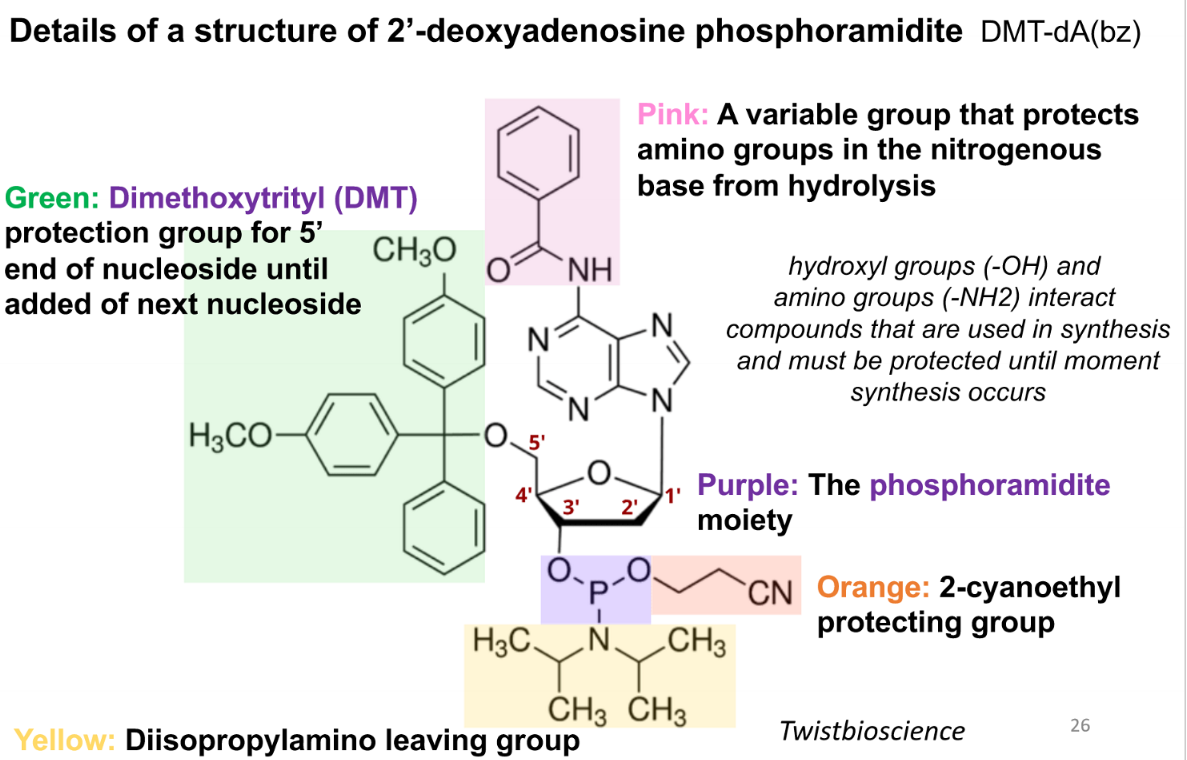
purpose of the pink group in a phospoamidite
variable group that protects amino groups in the nitrogenous base from hydrolysis
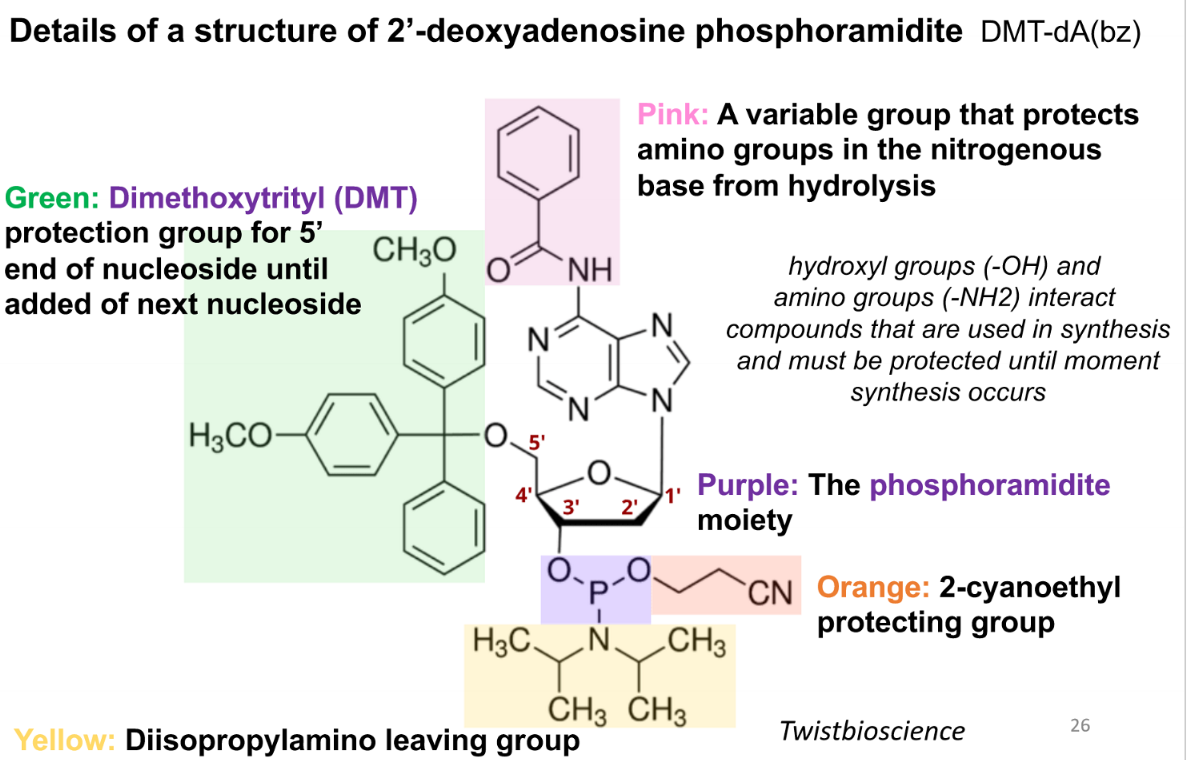
purpose of green (DMT) group in a phosphoamidite
protection group for 5’ end of nucleoside until addition of next nucleoside
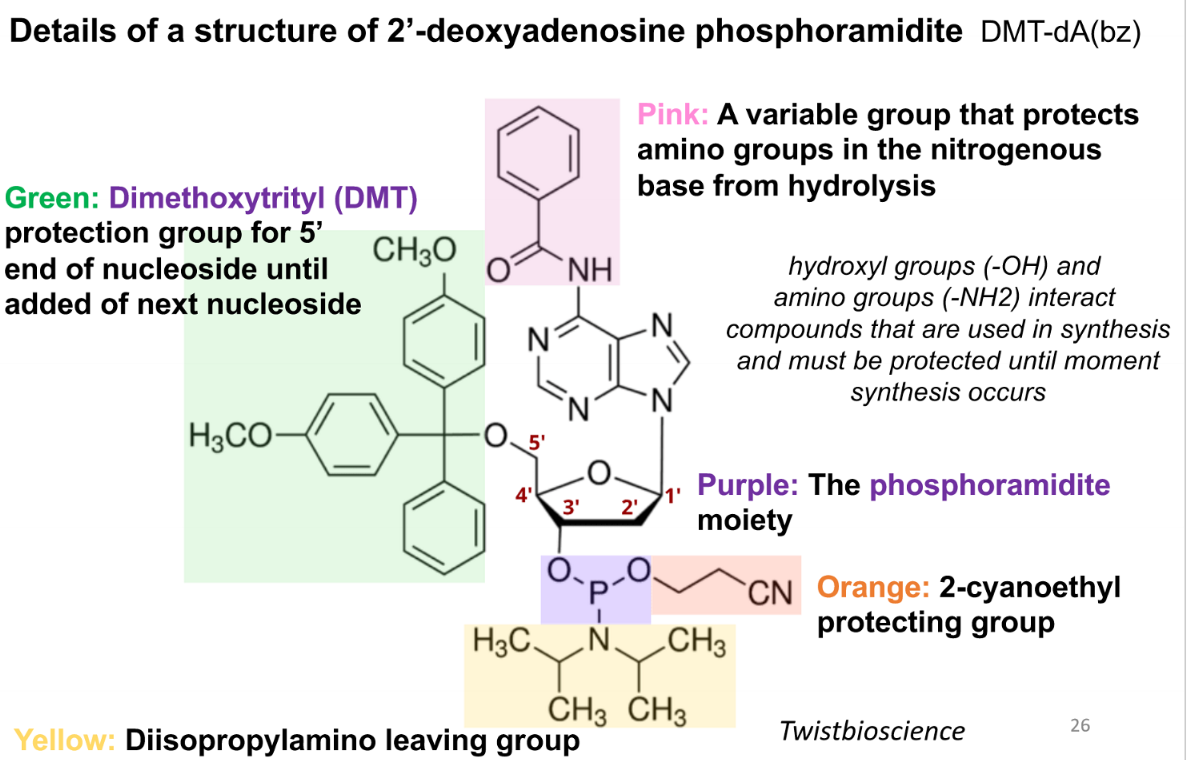
purpose of the purple group in a phosphoramidite
the phosphoramidite moiety
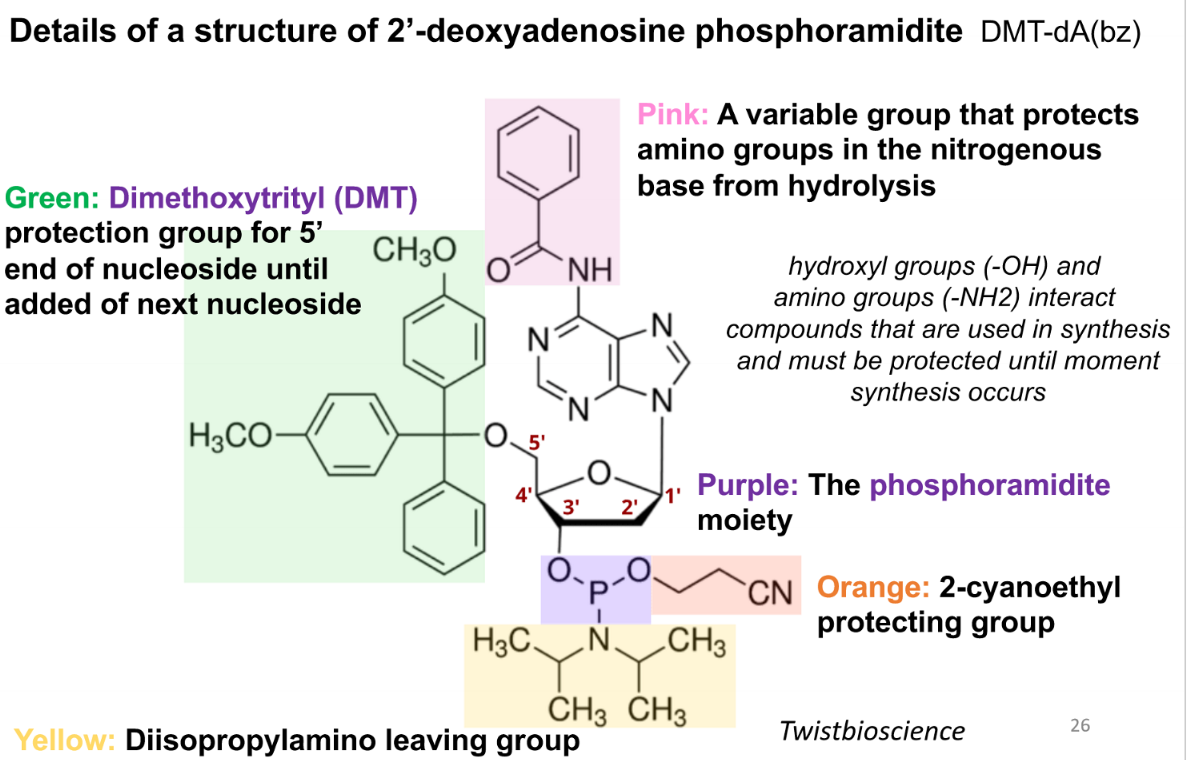
prupose of the orange group in the phsophoramide
2-cyanoethyl protecting group
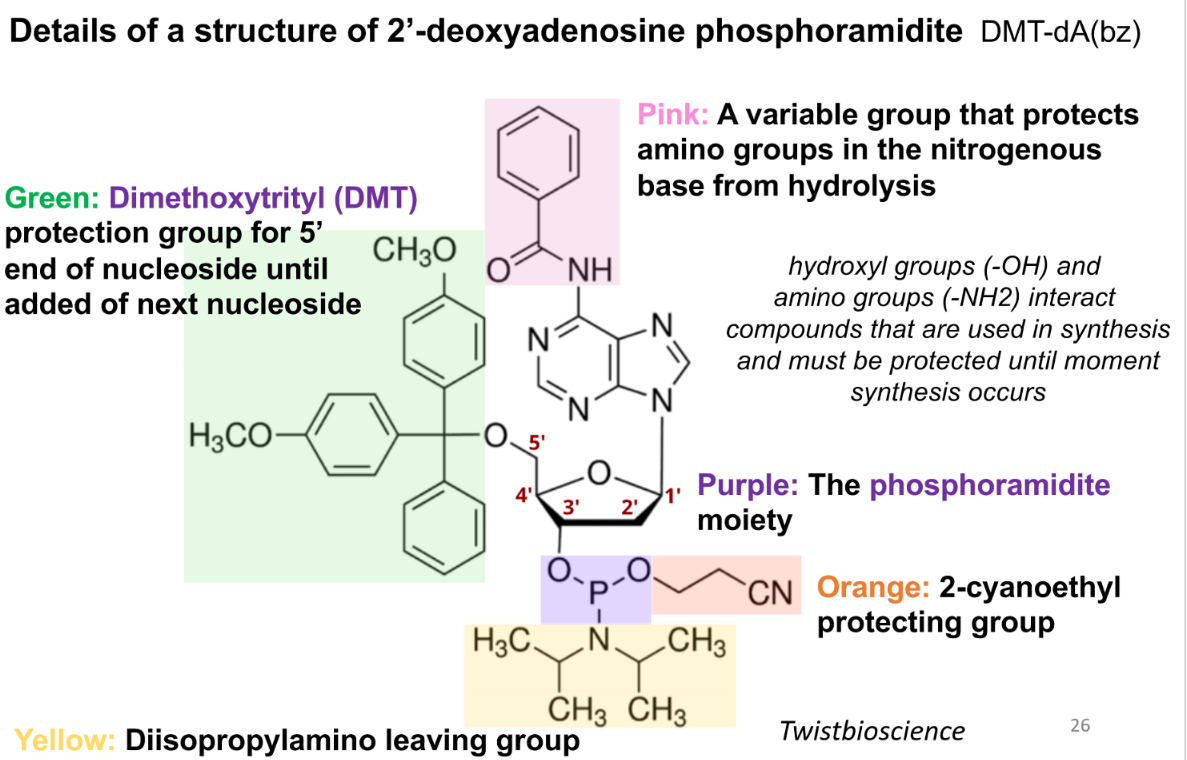
purpose of the yellow group in the phosphoamidite
diisopropylamino leaving group
steps of nucleic acid synthesis by phosphonamidite
attach 3’ end of the protected nucleotide to a support (silica)
remove the 5’ DMT protecting group
add next protected nucleoside, which is a nucleoside phosphoramidite
oxidize backbone linkage
repeat steps 2-4
remove protecting groups
cleave from silica support
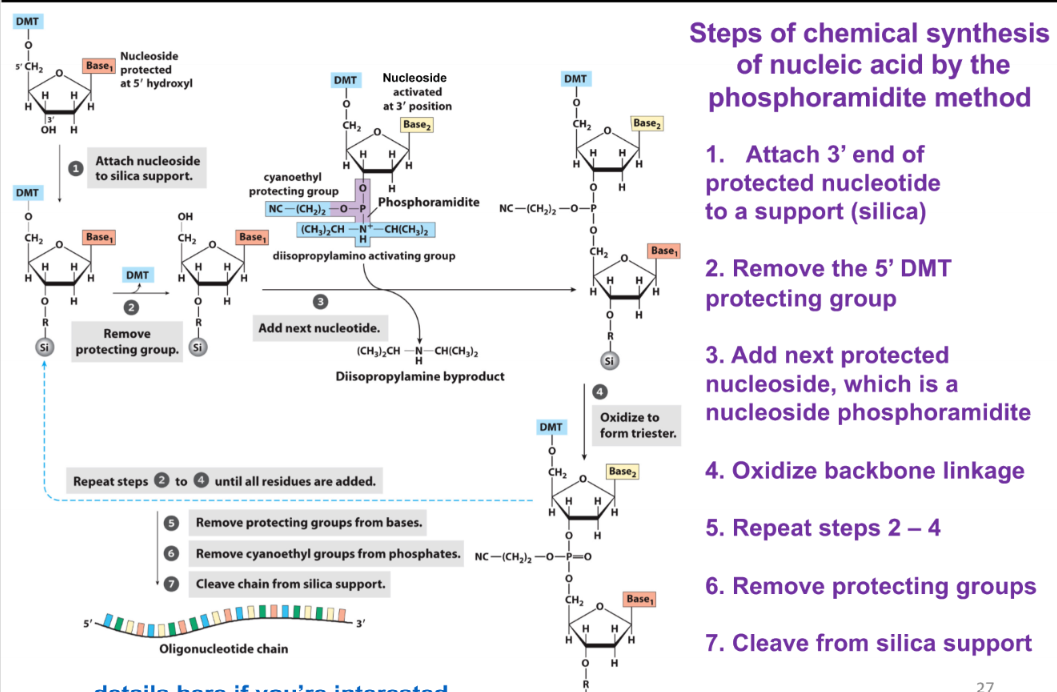
chemical synthesis of nucleic acids by the phosphoramidite method proceed in what direction?
3’ → 5’ (opposite of biological synthesis of nucleic acids)
DNA sequencing
process of determining a DNA sequence
what size bp fragments are analyzed in first generation DNA sequencing
500-1000 bp fragments
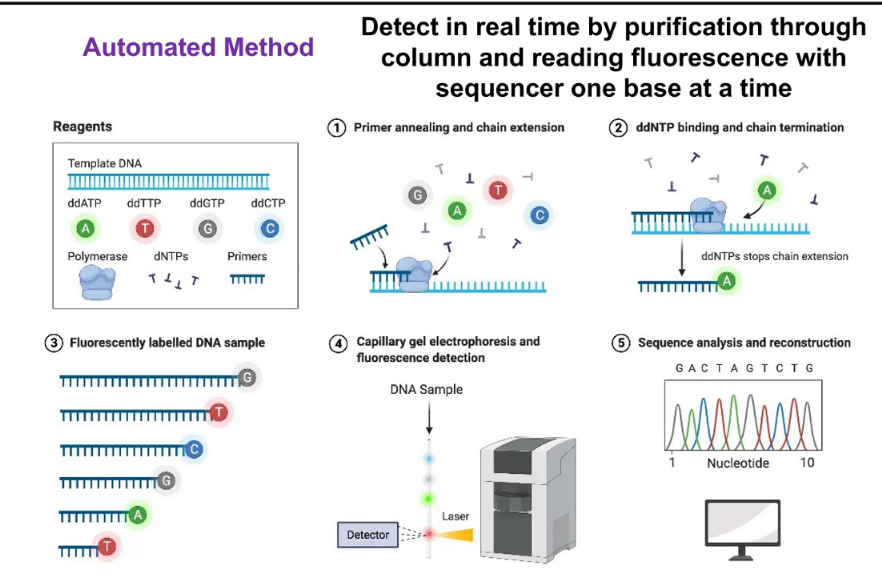
Sanger sequencing
based on random incorporation of chain-terminating dideoxynucelotides during in vitro DNA replication
ingredients for Sanger DNA
dideoxy or chain-terminating version of all four nucleotides (ddNTPs) each labeled with a different fluorescent dye or radiolabel
thermostable DNA polymerase
Buffer
primers (usually DNA)
dNTPs
template to be sequenced
why are ddNTPs chain terminating?
because they inhibit elongation by DNA pol
manual method of Sanger
separate the DNA fragments using electrophoresis to determine each nucleotide in sequence
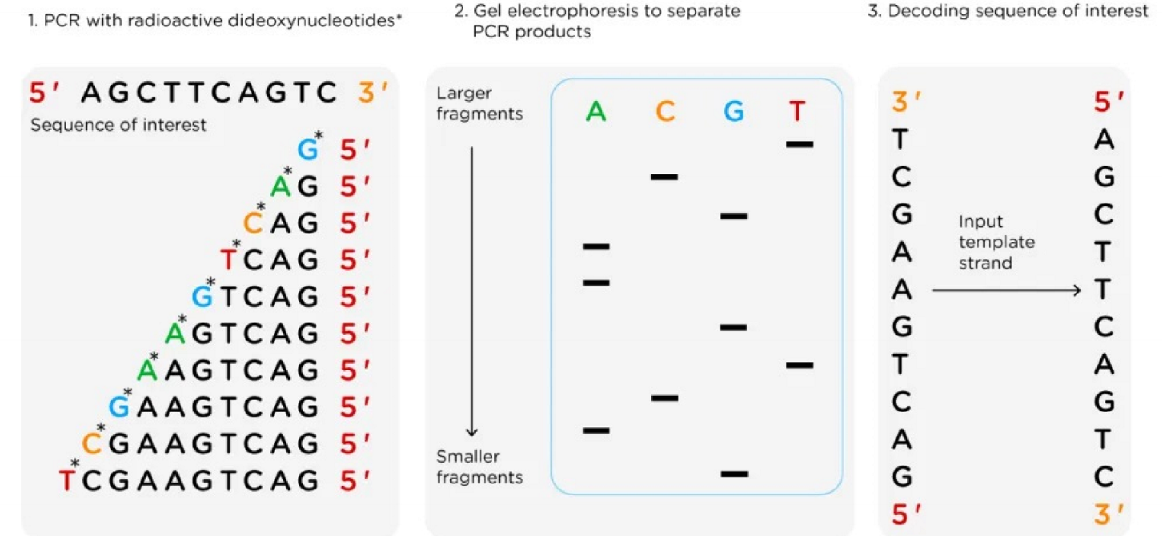
automated method of Sanger
detect in real time by purification through column and reading fluorescence with sequencer one base at a time