DNA Repair mechanisms part 1 & 2
1/23
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
24 Terms
Mechanism of reversal DNA damage
Photoreactivation
Alkyl transferases
Mechanism of excision repair
Base excision repair (BER)
Nucleotide excision repair (NER)
Mismatch repair (MMR)
Mechanisms of strand break repair
Single strand break repair
Double strand break repair
Mechanisms of tolerance of DNA damage
Replicative bypass
Trans-lesion DNA synthesis
Damage reversal: simple processes involving a single gene product (protein)
photoreaction using DNA photolyases
dark reaction and light reaction
involve DNA photolyase that switch out the thymine and then add on the adenine
The result is the removal of thymine dimer and success A-T binding restore
Damage reversal: simple processes involving a single gene product: reversal of monoalkylation - O alkylated base damage
enzyme: O6-methylguanine methyltransferase (MGMT)
O6-methylguanine binds with T
it is a suicide enzyme as its cystine end binds to the methyl on the O6
so in a tumour that is treated with TMZ, the MGMT would reverse the alkylating damage which desensitise the tumour to TMZ.
What are AlkB homolgues (ABH2,3, etc…?)
Members of the 2-oxoglutarate and oxygen dependent dioxygenases
Also has a role in reverse alkylation which turns 1-methyl adenine to adenine
Another enzyme that causes chemoresistance to TMZ
Excision of DNA damage: Base excision repair (BER) - What are the components?
DNA glycosylases - diffuse along the DNA to look for damaged bases, kink out the DNA, flip out the modified nucleotides into a pocket and cut the N-glycosidic bond. The glycosylase remains associated at the abasic site until displaced by APE 1
APE1 endonuclease - cuts the phosphodiester back bond to generate a single strand break
polymerase b: new DNA sythesis which uses the undamaged template strand to insert the correct base
ligase 3: everything is sealed back in place by the ligase
Which DN glycosylases are involved in the BER of 8-OxoG?
What happens in MUTYH mutation?
OGG1 - removes O opposite C
MUTYH: removes A opposite O
MUTYH mutation: responsble for MUTYH assoicated polypopsis- which has high incidence of colon cancer
Result in G to T transversions eg. in APC gene - commonly mutated in cancer
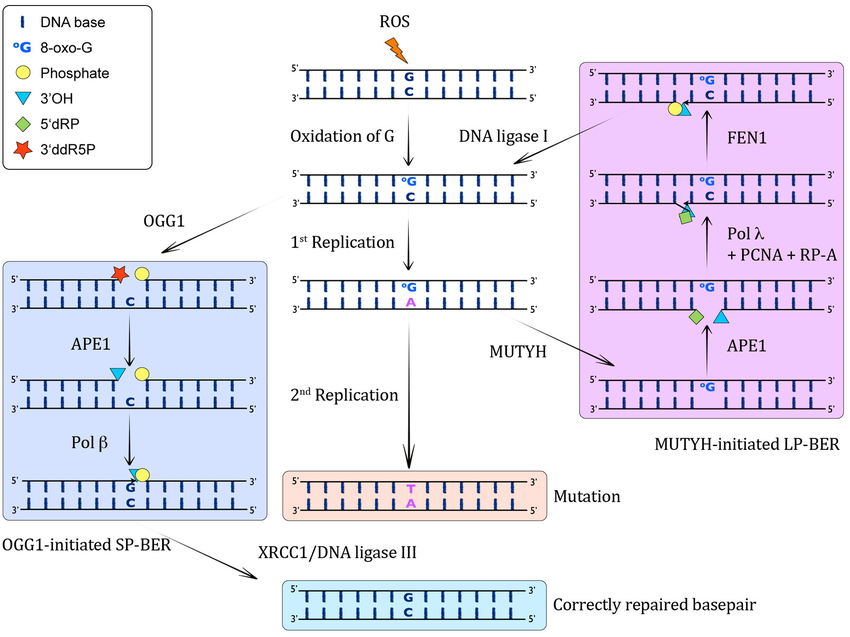
Unlike the DNA polymerases involved in replication, which insert A at O, the polB for BER preferentially …
inserts C opposite O
What is the role of Nucleotide excision repair?
Xeroderma pigmentosum - defective in NER
Recognising large lesions, covalent modification of DNA that distort the double-helix structure such as
UV primidine dimers
Bulky DNA products such as DNA cross link from bifunctional alkylating agents
Different in BER as it involves a dual-incision made on either sides of the lesion resulting in a removal of an 24-32 residue oligonucleotide (rather than just 1)
Xeroderma pigmentosum - skin tumour, extreme photosensitive skin
Steps in nucleotide excision repair
Recognition of helix-distorting lesions by XPC/ XPE
TFIIH complex (with XPB and XPD helicases) are recruited to open the bubble and unwind 20-30 nucleotides around the lesion - repair bubble
The lesion induces the blockage of XPB and XPD progression and stop it from advancing smoothly across the strand, which verify the damage and recruit nucleases to cut the 3’ and 5’ sides of the lesion. XPA and RPA stabilise the open complex.
End result - release of damaged DNA in an oligonucleotide
Repair synthesis by polymerase and synthesis of final phosphodiester bond by DNA Ligase 1
Transcription coupled repair is also similar to this
Low activity of … in testicular cancers appears to be responsible in part for sensitivity if thse tumours to cisplastin
XPA
Mismatch repair
Occur right after replication, works after DNA synthesis.
MutS and MutL in bacteria - MSH2-6 heterodimer and MSH1-3 heterodimer in human
In human, strand discrimination relies on pre-existing nicks in the new strands (Okazaki fragment junctions, PCNA and RFC signal at a gap)
The repair complex introduces a nick on the new strand near the mismatch
MutS/Mut L complex act as a sliding clamp along either direction of DNA, when encounter PCNA and RFC at a gap, RFC is displaced and allow the EXO1 to bind to the clamp and excise the mismatch. Polymerase + ligase close the gap.
How does MMR confers sensitivity to some DNA damaging agent and what are they?
DNA replication in the presence of modified base (eg O6-MeG), leads to misincoporation of T which stimulates MMR activity, However, it can only repair the T on the daughter strand which generates a futile cycle, which persistence recruitment of MutS MutL can upregulate ATM/ATR → Chk2 inducing apoptosis
or replication → mutation
What is the cause of hereditary nonpolyposis colon cancer - Lynch syndrome
due to mutation in MMR genes
Germ line heterozygotes + LOH
High mutation rates in MMR-deficient tumour leads to high density of no antigens and high CD8 T cells infiltration
Pembrolizumab - antibody that blocks PD-1
Single strand break repair - what is it and which is the important molecule? which drug is it inhibited by?
Arise during the final stages of BER, where they are rejoined by ligase 3
Can also be generated by DNA-damaging agents such as ionising radiation ,
Important as unrepaired breaks lad to collapse of replication fork resulting in formation of double strand break
PARP-1 is the important molecule
it catalyses NAD+ to poly (ADP-ribose) to throw an “emergency flare” at the DNA damage site for recruitment of repair
PARP-1 binding to ssb recruits ligase 3, DNA pol b and suppresses unwanted recombination
10^4 spontaneous ssb occur in each cell everyday. In the absence of PARP-1, ssb can be converted to dsb due to stalling of replication fork. PARP-1 hence is essential for survival if HRR is defective
Both PARP-1 and HRR defective? What drug can we use?
Synthetic lethality: combination of two genetic events results in cell death
Olaparib
Very good with BRCA1 -/- mutant in breast cancer
Types of DNA double strand break repair
NHEJ - error prone
MMEJ - error prone
Single strand annealing - error prone
HRR - high fidelity
NHEJ
Ku70/Ku80 heterodimer formation - binds to the broken DNA ends
Recruiments of the large scaffold DNA-Pkcs
Recruitment of low specificity Ligase IV
Ligase IV knockout mice are non-viable
Knock out of DNA-PK gives viable mice but lack of VDJ rejoining lymphocytes
HRR pathway key proteins
MRN complex - Rad50, NBS, Mre11
Rad52, 53
BRCA1 and BRCA2
ATM exists as … that is activated by autophosphorylation in response to DNA …
ATM exists as an inactive dimer that is activated by autophosphorylation in response to DNA double strand break
Tolerance of DNA damage mechanism
1) Gap formation and recombination
In this case, a gap is formed opposite the lesion, which is subsequently repaired by a homologous recombination mechanism
2) Translesion DNA synthesis by the error prone DNA polymerase (eg, pol-k ) that can replicate past the unpair regions or make sensible base selection?
The MRN complex is a sensor that detects DNA double strand breaks and initiates Homologous Recombination Repair (HRR). What role does MRE11 play in this process?
Recruits the RAD52 helicase