AMT Final
1/484
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
485 Terms
What is hybridization
Hybridization = the binding of a single-stranded molecule of DNA to a complementary single-stranded target DNA molecule
- “sticking together” or “anneal”
- Occurs between PCR primers and the target template
- Usually used to describe binding of special oligonucleotides/probes
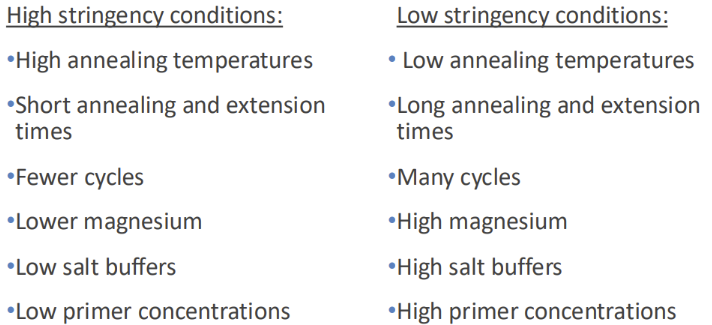
What is stringency
Stringency = the combination of conditions under which the target is exposed to the probe
- Conditions of HIGH stringency are more demanding of probe binding (more specific)
- Conditions of LOW stringency are more forgiving (less specific)
What factors affect the stringency of a probe binding to its target
1. Temperature = as temp INCREASES, stringency INCREASES (hotter = only [erfect matches can stay bound)
- Creates a more challenging environment for the probe to bind, only allowing perfectly-matched probes to hybridize
- If temp is too low, the probe will find less-than perfectly matched sequences to bind
- If temp is too high, the probe cannot form its bond and will fail to hybridize
2. Salt concentration = as salt conc. Increases, stringency DECREASES
- Because salt promotes DNA binding (stabilizes DNA) it promotes any kind of binding, even non-specific kinds
- By lowering salt, only the most specific probe target hybrids can stay stable
- Too little salt means no hybridization at all
3. Denaturant in buffer = INCREASE denaturing/destabilizing agents will INCREASE stringency
- Agents = formamide, urea, TMAC (all break up secondary structures)
- Presence of formamide increases stringency because it promotes denaturation even at lower temps (makes it harder for probe to bind => more specific binding)
- When the probe and target are prone to denaturation, only the most specific probe-target hybridization can occur
4. Time
a. Hybridization time = INCREASED time of hybridization (incubation of probe and DNA) DECREASES stringency
- The longer the probe mixes with the DNA, the more likely it will bind non-specifically
b. Wash time = INCREASED time of washing the blot INCREASES stringency
- The longer the wash time, the more likely it will detach weak non-specific bonds, leaving only specific probe target complexes (washing too long can knock off the targets)
5. Length of probe = a LONGER probe will bind under MORE stringent conditions
6. GC content of probe = HIGHER GC probe will bind under MORE stringent conditions
- Presence of formamide will allow GC-rich probes to denature and hybridize at lower temperatures than without formamide
Conditions that are not stringent may be more likely to produce non-specific products for a GC-rich probe than for an AT-rich probe
What is a probe
Probe = a single-stranded fragment of nucleic acid attached to a signal-producing moiety
- Used in molecular methods to identify a sequence of interest within a large mixture of nucleic acid
- Hybridizes specifically with the target DNA/RNA to be analyzed
- Can be denatured DNA/RNA
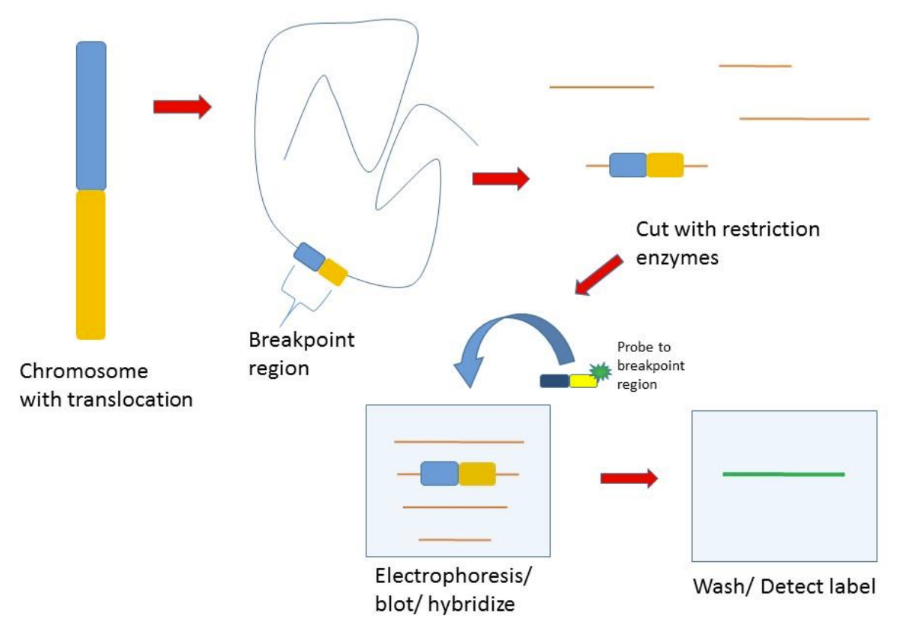
How does the size of the probe determine hybridization
DNA probes:
Size of the probe determines hybridization
- The length of the probe helps determine the specificity of the hybridization reaction
- When probing the entire genome, longer probes are more specific because they must match a longer sequence on the target (unlikely to be found elsewhere in the genome)
o Shorter probes are more likely to be found in multiple locations in the genome
§ Because higher chance to find more identical sequences
- Short probes play an important role in mutational analysis, often with PCR amplification, because shorter probes can be sensitive to single-base mismatches
o Probes will only bind if there is mutation
What are probe labels
Probe labeling: the probe must be labeled and generate a detectable signal (how to detect if probe has bound or not)
- Classically = radioactive labels like 32P
o Introduce nucleotides containing radioactive phosphorus into the probe
- Common = non-radioactive methods (exclusively used in medical laboratories)
o Usually fluorescently tagged
How does the length and location of the probe affect binding/PCR
Probe design
- Length of probe determines specificity (depending on application)
o Genomic DNA (no PCR) = long probes are more specific
o PCR amplified DNA (smaller pieces of DNA) = short probes are better to find mutations
- Location/sequence of the probe can affect binding performance
o Internal complementary sequences will fold and hybridize with itself rather than staying single-stranded and linear
What is the relationship between stringency and PCR
The conditions of PCR determine stringency of PRIMER binding and thus determine the specificity of the PCR reaction (do you produce your target band and ONLY your target band)
IMAGE: Don't need to make all these changes at once. If you had many non-specific products start on the left or if you had no products start on the right
The conditions you use for probe incubation determine stringency of hybridization
What do different probe results mean (in relation to stringency)
1. If conditions make it TOO HARD for the probe bind, it won’t bind and send fluorescent signal EVEN IF the mutation is present
- Stringency too high
2. If conditions make it TOO EASY for the probe to bind, it will bind and send fluorescent signal EVEN IF the mutation is NOT present
- Stringency is too low
3. Need conditions for probe binding (stringency) to be JUST RIGHT to produce specific probe binding when the mutation is present, and no probe binding when there is no mutation
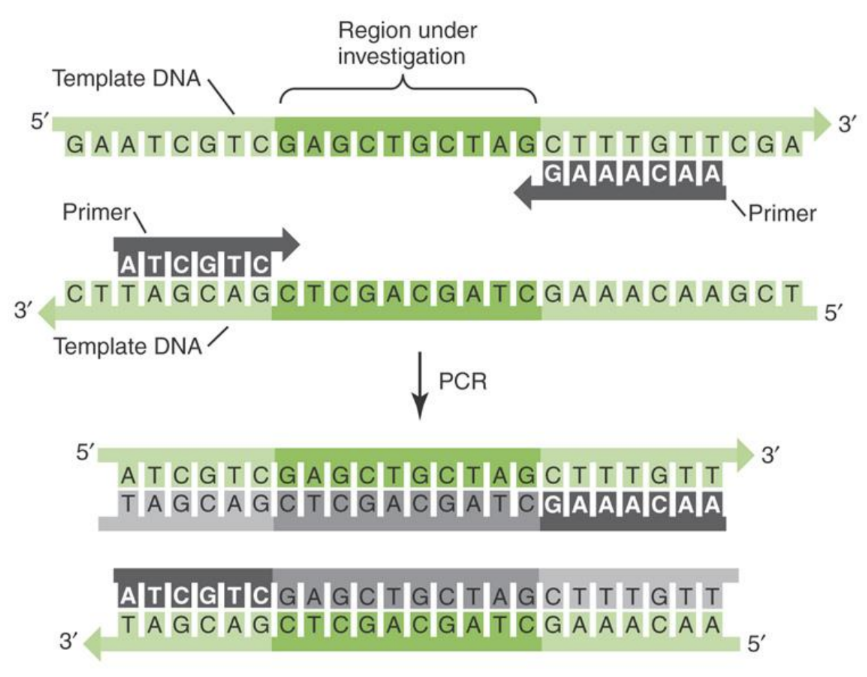
What is a primer
Primers = determine the specificity of the PCR reaction (small oligonucleotides)
- 15-35 bases long
- Complementary to sequences flanking the region to be amplified
- Single stranded and used in pairs
How do primers bind
How primers bind:
- Binding of primers is determined by the sequence, buffer conditions, and temperature
- Ideal binding/hybridization of primers can be estimated by calculating the melting temperature
What are the principles to primer design
- Avoid primers that anneal to themselves or other primers
o Avoid complementarity at the 3’ ends
- Choose primers specific to the target sequence
o Avoid simple repeat sequences that are common across the genome
o Beware of related genes, which may have conserved sequences
- Primer size is optimum between 18-25 bases in length
o Primers smaller than 15 bases will likely find non-specific binding sites
o Primers bigger than 30 bases will not bind efficiently
- Match primer Tm’s closely
o Within a few degrees of each other
- For standard, end point PCR, choose a product length ~200-500 bases
o Big enough to see on gel
o Small enough to amplify efficiently
- GC content should be ~50-60%
- Avoid long stretches of any one nucleotide
o PolyA runs or polyT runs can cause breathing or transient opening of primer target complex and mispriming
o Poly G or PolyC runs can cause mispriming
- A few G or Cs on the 3’ end is preferred
o GC clamp = reduces breathing, increases priming efficiency/yield
Where do primers bind on a DNA sequence
Primers flank to the region of interest (complementary to sequence)
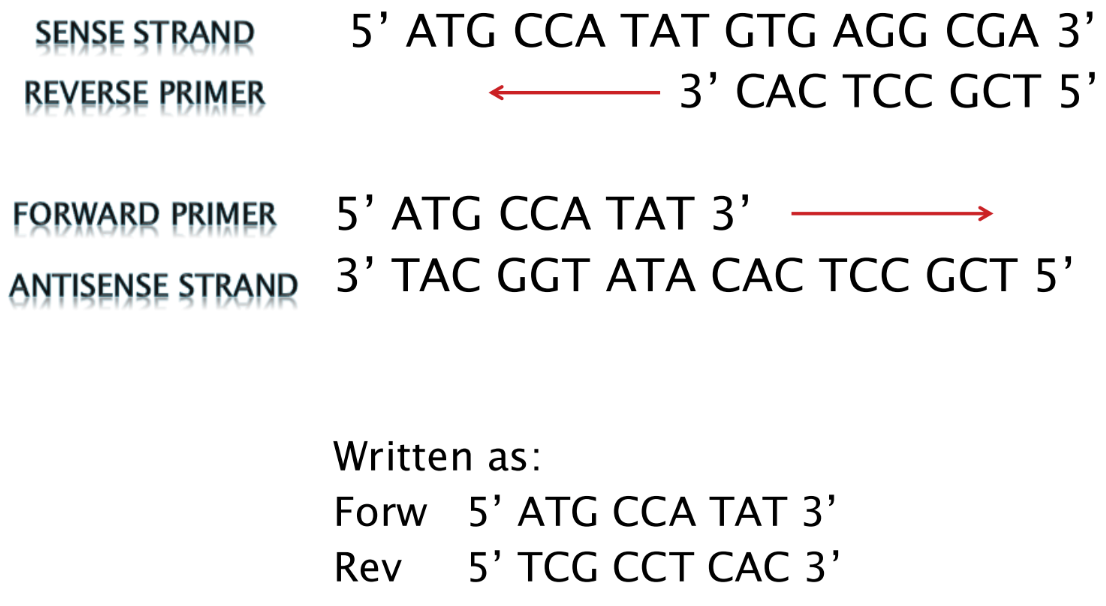
What is a forward primer
Forward primer = binds to the antisense strand
- Sequence is the SAME as the sense strand (but binds to antisense strand)
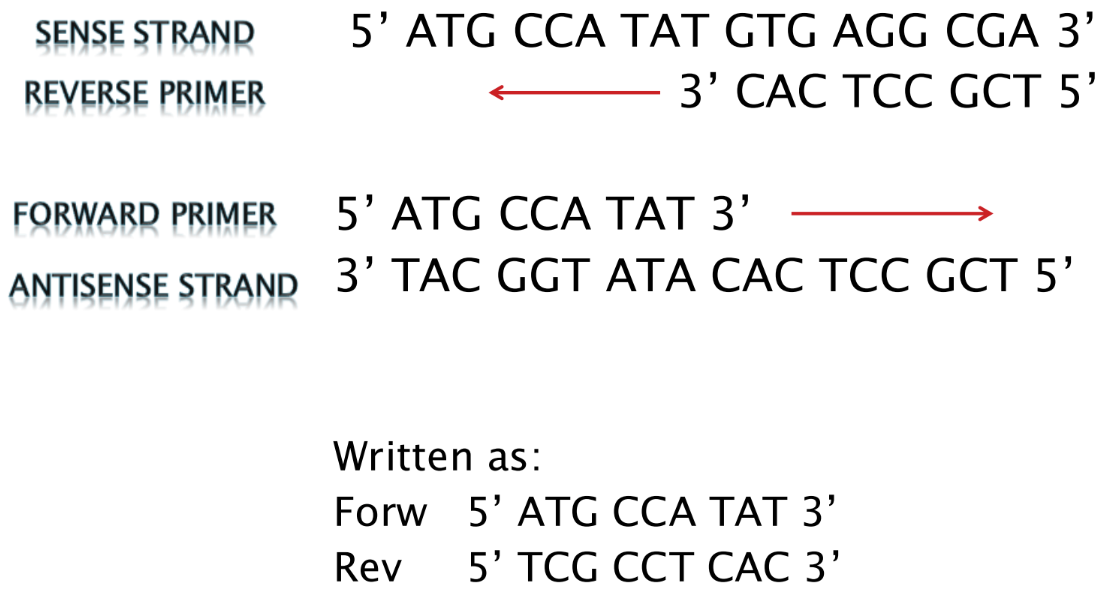
What is a reverse primer
Reverse primer = binds to the sense strand, downstream
- Sequence is the SAME as the complement of the sense strand (but binds to sense strand)
o Written 5’ to 3’, making it antiparallel to the sense strand sequence
Predict the amplicon size produced by PCR
Size of PCR product = determined by the distance between the forward and reverse primers
o Far away flanking = bigger product
o Close flanking = smaller product
- Therefore, should be able to predict how the result should look like on a gel
Calculate the Tm’s of a primer
look at pic
need to know (WILL ASK)
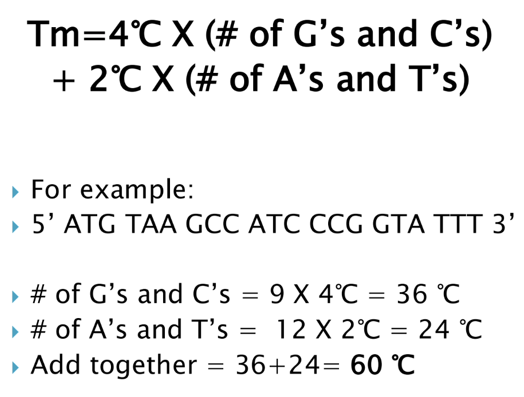
What is melting temp. (Tm)
Melting temperature (Tm) = the amount of energy required to separate the primer from the template (target) strand (how well primers remain bound)
- Can be used to estimate ideal primer binding/hybridization
- At Tm, half the sequence is double stranded (primer bound to target) and half is single stranded (primer separated from target)
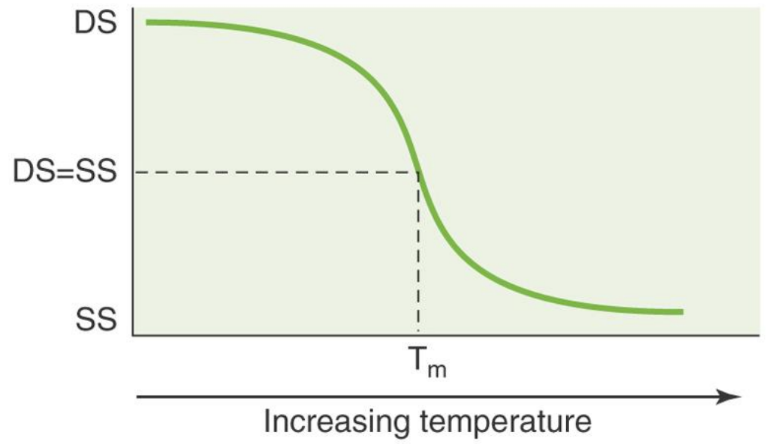
How can Tm’s be adjusted
Adjusting Tm’s
- Tm can be increased by increasing the length of primers
- Primers with more G/Cs will have higher Tms because they have more Hydrogen bonds
- Tm's of the primer pair should ideally be within 2-3 C of each other
Determine an optimum annealing temperature for a given primer pair
Estimating annealing temperature (Ta) = use an annealing temp 5 C lower than the lowest Tm of the primer pair
- Starting point for optimization (ensures some products will be produced)
- Primers should anneal to the template before the template anneals to itself (ideally)
Annealing temp is too low:
- One or both primers may anneal to sequence other than the target
o Partial annealing, single base mismatches may be tolerated
- Non-specific amplifications, reduced yield of product
Annealing temp too high:
- Temp may be too high to allow primer binding
- Reduced to no product yield
Optimum annealing temp:
- Single product is formed, high intensity band on gel, no other bands or smearing
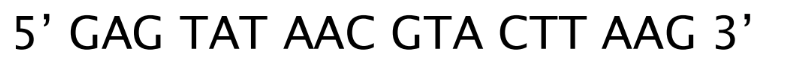
Design primers by hand for a given target sequence
Forward primer = 5’ GAG TAT 3’
Reverse primer = 5’ CTT AAG 3’
List the components of a PCR reaction with optimum concentration ranges
Buffer (MgCl2, KCl, & Tris) = 10X stock diluted to 1X
MgCl2 = 1-4mM final concentration
dNTPs = 0.1-0.5 mM each – 0.2mM (200uM) each is common)
Primers = 100nM to 1uM each (200nM/0.2uM is typical for each primer)
- Stock concentration = 0.2-100uM
- Should be in excess in the reaction so the DNA doesn’t bind back with each other
Taq polymerase = 0.5-2.5 Units/50uL reaction
- 1-2.5 Units/50 ul reaction)
- LOWEST VOLUME (critical to pipette into liquid)
DNA template = 1ng-1ug of DNA per reaction (10ng first time)
- 1-100ng DNA/reaction
Describe typical PCR conditions including times and temperatures
Temperatures/times:
Initial denature step = ~93-95 C for 30 sec to 10 min
25-40 cycles of…
Denature = ~94-96 C, 15-60 sec
Anneal = ~50-70 C, 5-90 sec
Extend = ~68-72 C, 5-60 sec
Final extension = ~68-72, 2-10 min
Hold at 4 C
What is HOT-start PCR
HOT-start PCR (taq will only be activated when it’s heated, therefore, it’s ok to leave at room temp)
- If Taq polymerase is allowed to activate before cycling starts, competing side-reactions in your PCR tube can occur during pre-PCR setup
o Because standard Taq polymerase has some activity at 25C (room temp)
- Prep samples on ice (cold-block) to prevent Taq activity until it’s time to start PCR
o Also protects the enzyme from degradation over extended periods
- Most modern manufactures make heat-activated Taq polymerase
o There's an antibody or other chemical modification attached to the Taq enzyme that prevents Taq activity until it reaches several minutes at denaturation temps (~94C)
What changes must you make to a master mix to optimize it
- Buffer components
o Supports the polymerase enzyme
o Includes salts
§ Salts in buffer affect denaturing and annealing temperatures and polymerase activity
§ Stabilizes DNA (harder to denature during PCR)
Increasing salt...
· Slows denaturation of long DNA products
o Can lead to long non-specific products
· Preferential amplification of short products
o May or may not include MgCl2
§ Too little magnesium
· Lowers enzyme efficiency
· Low yield of PCR product
§ Too much magnesium
· Promotes misincorporation (wrong bases, errors introduced)
· More non-specific products
§ If you need high fidelity, stay on low end of Mg conc.
§ If you aren’t getting product, increase Mg
(the image Shows how Magnesium concentration affects the product)
o Chelators
§ EDTA can chelate the magnesium (holding it and prevents it from being used in reaction)
· If EDTA present, may need to increase magnesium in reaction (because it can lower the amount of magnesium)
· EDTA used to support DNA (keeps it safe)
o Other potential buffer components
§ BSA = general protein, enzyme stabilizer
§ DTT = reducing agent, enhances enzyme (to a point) (enhances to make it easier to denature)
§ Formamide = denaturant, lowers denaturing temp
· Used with high secondary structure (GC rich DNA)
§ Other co-solvents
· TX-100, glycerol, and DMSO = reduce secondary structure
· Used for GC rich templates
- PCR additives
- Primer concentrations
o Too high of primer concentrations encourages mis-priming
§ More non-specific products
§ More primer dimer
o Long reactions may need more primers to prevent exhaustion of reagents
o Fewer primers = more specific product
- DNA template
o Too little template = little to no product, more primer dimer
o Low (but not too low) template = increases specificity
o Too much template = mis-priming, non-specific products
- Taq polymerase (not the go to in optimization)
o Keep on ice
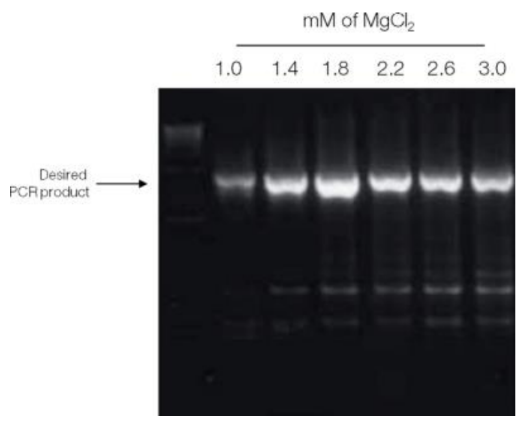
Predict the consequences of changes in PCR master mix and thermocycling conditions
Optimizing thermocycling
- Denaturation (94-96)
o Not typically necessary to adjust, unless its a GC rich template
o Initial denaturation step at ~95C for 2 mins is recommended prior to cycling to fully denature DNA
§ Initial denaturation may also be used to activate “hot-start” Taq polymerases (critical step)
- Annealing (dependent on primer Tm’s)
o Increase annealing temp if there are non-specific products
o Shorten annealing time if there are non-specific products
o Decrease annealing temp if there is no product (although there could be other reasons for no product, like forgetting ingredients)
o Annealing is more specific at shorter times, some reactions can anneal in just a few seconds
§ 15-30 sec annealing times usually adequate
§ Less opportunity for primers to bind to wrong thing (get in “trouble”)
- Extension
o Decrease extension times to promote shorter products or to eliminate larger non-specific products
§ 15 sec extension time may be more than sufficient for < 500bp
o 1 min for 1kb of product
§ Some polymerases are much faster, so may need less time
- Cycle number
o Too few cycles = less product
o Too many cycles = non-specific products
o Increase cycles if you have very little template
o Decrease cycles if you have a lot of template (or decrease template)
o Can be optimized by creating a series of identical reactions and then removing them one at a time from the thermocycler at different cycles
§ Analyze how target template yield and specificity changes at different cycle numbers
What is the goal of PCR
Goal = a STRONG (intense) target band when loading even a small aliquot of the PCR reaction on a gel, with minimal to no non-specific products
What should you do your PCR has little to no target band
For little to no target band:
1. Check your math
2. Verify all components are present
3. Verify primer design
4. Consider if DNA sample has inhibitors (amplified successfully in other reactions?)
5. Lower annealing temperatures
6. Add more template or cycles
7. Add more magnesium
8. Use PCR enhancers or higher denaturation temps for GC-rich targets
What should you do if your PCR has non-specific products
For non-specific products:
1. Increase annealing temperature
2. Reduce primer concentrations (especially if there are abundant primer dimer complexes)
3. Shorten annealing and extension times
4. Reduce cycles or template
5. Reduce magnesium
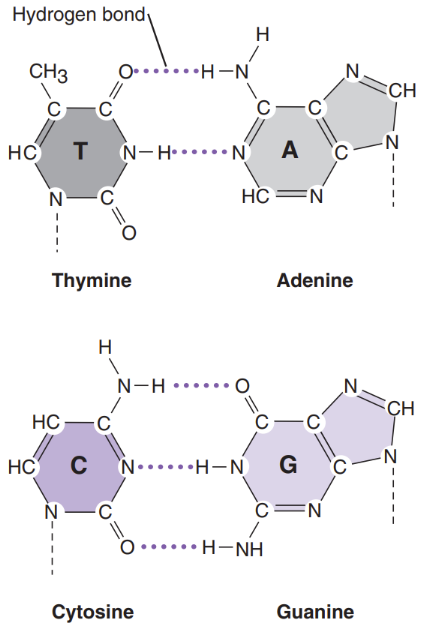
What is a nitrogen base
Nitrogen bases = attached to deoxyribose sugar. Are the 4 building blocks of life
Adenine
Cytosine
Guanine
Thymine
Purines = bases with a double-ring structure
Ex: G & A
Pyrimidines = bases with a single ring structure
Ex: C & T
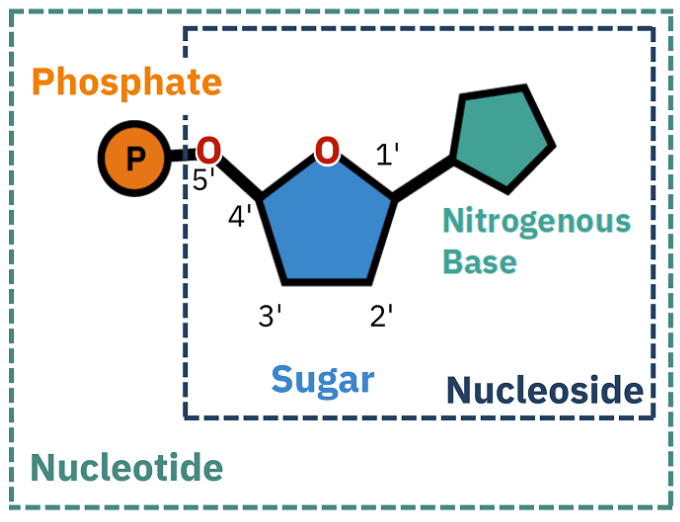
What is a nucleoside
Nucleosides = A nitrogen base bound to an unphosphorylated sugar
When the ribose sugar is phosphorylated...
Mono = nucleoside
Ex: Adenosine monophosphate (AMP)
Triphosphate = nucleotide
Ex: Adenosine triphosphate (ATP)
di-phosphate = 2 phosphorylation
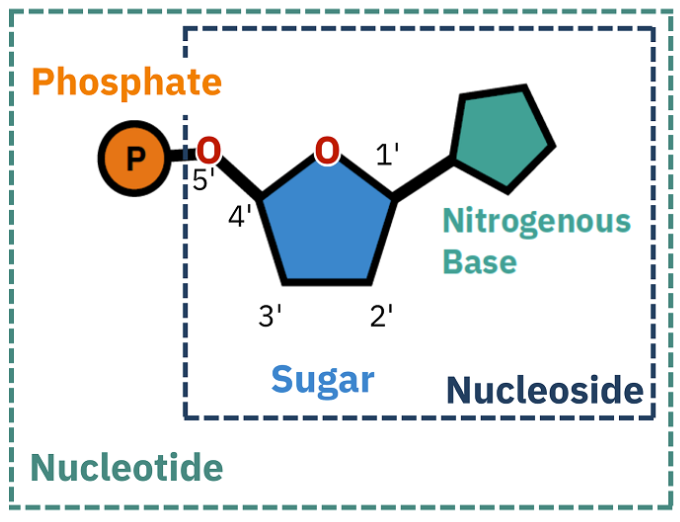
What is a nucleotide?
Nucleotides = essential building blocks of DNA and RNA, composed of a nitrogenous base, a five-carbon sugar (ribose or deoxyribose), and a phosphate group
What is a nucleic acid? What is it’s structure
Nitrogen bases attached to a deoxyribose sugar form a polymer with the other deoxyribose sugars of other nucleotides via phosphodiester bonds
Nucleic acid = a macromolecule made of nucleotides bound together by the phosphate and hydroxyl groups on their sugars
Grows by the attachment of 5’ phosphate group of an incoming nucleotide to the 3’ hydroxyl group of the last nucleotide on a growing chain
Gives the chain polarity (5’ & 3’ end)
Hybridization = formation of hydrogen bonds between 2 complementary strands of DNA
What are the steps of DNA replication
Unwind the DNA via helicase
Primase adds the primer
DNA elongation via DNA polymerase
Makes leading and lagging strand
DNA ligase seals nicks and joins strands
What is polymerase
Polymerase = responsible for polymerizing the nucleotide chains
Uses a guide/template strand to know what nucleotides to add to a chain
What is exonuclease
Exonuclease = degrade DNA from free 3’ hydroxyl or 5’ phosphate ends
Don't work on closed/circular DNA
protects the sequence of nucleotides
What is endonuclease
Endonuclease = break the sugar-phosphate backbone of DNA
What is ligase
Ligase = an enzyme that forms phosphodiester bonds between existing DNA strands
Catalyzes the formation of a phosphodiester bond between adjacent 3’ hydroxyl and 5’ phosphoryl nucleotide ends
What is nuclease
Nuclease = natural components of cellular lysates
Important to eliminate or inactivate when preparing nucleic acid specimens for clinical analysis
What is helicase
analysis
Helicase = unwinds and untangles DNA for replication
The release of DNA for transcription, replication, and recombination without tangling is brought about through cutting and re-closing of the DNA sugar-phosphate backbone
What is methyltransferase
Methyltransferase = catalyze the addition of methyl groups to nitrogen bases, usually adenines and cytosines in DNA strands
What is gel electrophoresis?
Electrophoresis = movement of molecules by an electric current through a matrix/gel
DNA is negatively charged (because of the phosphate backbone) so it moves towards the positive pole
DNA travels at speeds inversely related to its size
Big molecules go slower (don’t migrate far in gel)
Small molecules go faster (further in gel)
What are the principles of electrophoresis
Principles:
Determine method for separation
Type of gel/matrix
Concentration (%)
Running parameters (time/voltage)
Select molecular weight marker (ladder)
Loading
Prepare samples (loading dye)
Load wells/column & document loading order
Perform electrophoretic separation
Visualize and document results
Stain
Chemiluminescence/UV/fluorescence
What’s the difference between agarose and polyacrylamide gels
Agarose | Polyacrylamide |
Very safe material/easy to work with
Ran in a horizontal format Lower resolving power
Good for separating larger fragments (very porous) Made from seaweed & agar components Concentration used: 0.5-5%
| Components can be toxic Usually ran vertically Finer size resolution (small DNA) DNA sequencing, capillary electrophoresis (1 base pair difference) Use for separating small fragments (& single stranded DNA)
Protein electrophoresis (western blotting) Concentration used: 3.5-20% |
Gels are porous like a sponge (allowing DNA to squeeze through with the electric field/matrix sieve
The concentration of gel/buffer affects the resolution of fragments of different size ranges
What’s the difference in agarose and polyacrylamide gel prep.
Agarose prep | Polyacrylamide prep |
|
|
What are the buffers that can be used in electrophoresis
Buffers:
Carries the current and protects the samples during electrophoresis
Typically comes as 10X or 50X stock
Dilute to 1X for working solution
Tris acetate EDTA (TAE) = DNA moves faster, but buffering capacity is smaller
Tris borate EDTA (TBE) = better buffering capacity, DNA moves slower
What’s the difference between TAE and TBE
TAE | TBE |
|
|
* Both buffers can be used interchangeably for PCR/many molecular diagnostic applications
What is the purpose of a loading dye
Loading dye = Gives color to DNA for easier visualization
Makes DNA denser than water so it sinks to bottom of well (weighs down)
Has tracking dyes that separate during electrophoresis to indicate progress of electrophoresis
Allows for visualization while loading
What’s the purpose of nucleic acid stain and detection reagents
make DNA visible
What is the purpose of a molecular weight marker (ladder)
Molecular weight markers (ladders) = a concentrated control stock of DNA fragments of known size
Necessary for determining the actual size of the DNA bands in your finished gel
Included in every gel
What are the general type of equipment used for electrophoresis
UV light box (transilluminator)
Gel documentation systems
Well combs = used to make wells when casting gel
Microwave = used to heat agarose
Casting tray = used to make gel
Gel box = runs reaction
Gel power supply = powers reaction
What’s the difference between the 3 nucleic acid application detection systems (ie. gelred)
Ethidium bromide | SYBR Green | Gel red |
|
|
|
How do you Calculate a sample mixture for loading onto an agarose gel (DNA, loading dye, water)
From the total final desired volume, subtract the loading dye and water amount. From there, subtract the amount of DNA you will use
If you have the DNA conc. From the Nanodrop. And you want to use DNA that is Xng. You will divide the desired ng by the DNA conc. To get the volume of DNA to pipette in (ul).
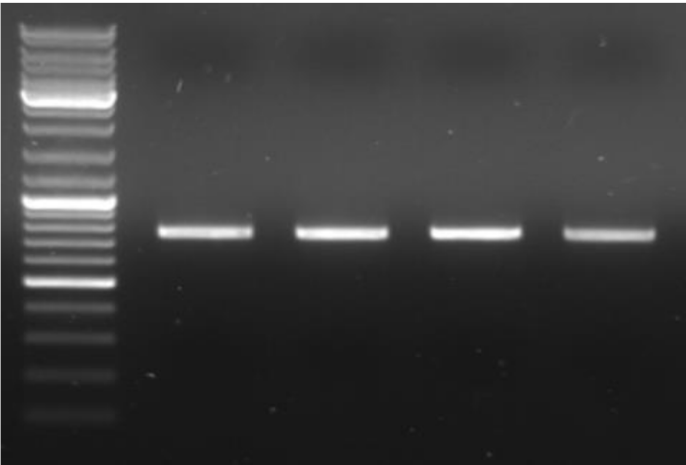
What is a problem with the shown image
No wells seen and no labeling
What is a restriction endonuclease
Restriction endonucleases = recognize specific short DNA sequences
Originate in nature (by bacterial cells as a defense mechanism against foreign DNA –phage)
Protect host by methylation of host DNA and cleavage of unmethylated DNA
Named after the bacteria it comes from
3 different types: Type I, II, III
Most are type II = cleave at specific recognition sites. Only unmethylated DNA
Recognize palindromes (in general)
Can cut 1 of 3 ways (sticky or blunt ends)
Type I: methylation/cleavage (3 subunits)
cuts >1,000 bp away from binding site
Ex: EcoAI
Type III: methylation/cleavage (2 subunits)
cuts 24-26 bp away from binding site
Ex: HinfIII
Can cut 1 of three ways: 5’, 3’overhang, or blunt end
Measured in units (U)
Check the compatibility of the enzyme with the buffer – not all buffers work with all enzymes (some enzymes can work with multiple buffers)
Use 10U of RE per microgram (ug) of DNA
They're named after the bacteria they come from
EcoRI = E. Coli (first one discovered)
Be aware of star acitvity
What is the molecular diagnostic use of restriction endonucleases?
Molecular diagnostic use = can see if there is some sort of mutation because a change in bp won’t allow for the RE to bind anymore (maybe a different RE will bind)
If there is a mutation, the RE will not cut (sample will look like control)
What' are the 3 types of ends an endonuclease can make
5’ overhang = sticky end because of anti-parallel nature
3’ overhang = sticky end because of anti-parallel nature
Blunt end endonuclease = leave no overhanging bases after separation because it is a palindromes
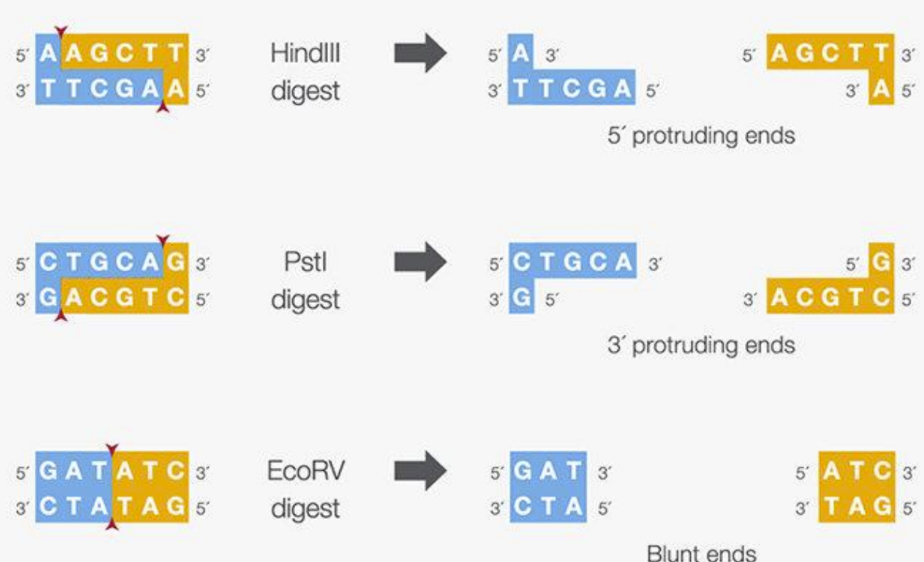
How do restriction enzymes cut DNA (#fragments)
Restriction enzymes
Recognize specific sequence (usually 4-6 nucleotides)
Cut the DNA by breaking the phosphodiester bond on both strands
Cutting results in 2 or more fragments
Smaller recognition sequences results in more fragments generated because it is easier to find a match for a short sequence
Resolve fragments by gel electrophoresis
* The number of times a specific sequence occurs in a given organism is approximated by...
Genome size in nucleotides/4^n
n = the length of the recognition sequence
* Master mix should be on cold block and gently mixed
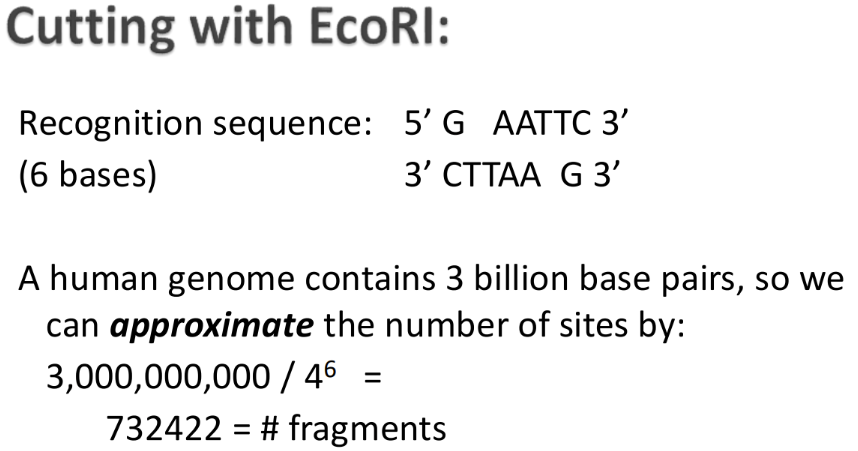
What is restriction enzyme mapping?
Restriction Enzyme Mapping: After digesting the DNA with RE and resolving the fragments by gel electrophoresis...
Number of bands indicates the number of restriction sites
Size of the bands indicates the distance between restriction sites
What are the detailed steps of restriction enzyme cutting process?
Detailed steps:
Consult the enzyme data sheet for details
It's important to find out the correct conditions for the enzyme that you’re using (usually provided by manufacturer)
Set up master mix on cold block
Mix gently by flicking, then briefly spin
For human genomic DNA = RE reactions typically incubate at 37°C for 5-18hrs
Enzymes often must be heat inactivated after reaction is completed
Usually between 55-88°C for 20 mins.
Analyze by gel electrophoresis
What is star activity
Star activity = When RE cuts the DNA too many times, results in extra bands (RE GONE NUTS)
Heat inactivation required to stop RE from over cutting
On a gel, there will not be a DNA smear visible at the top of the gel because DNA is degraded
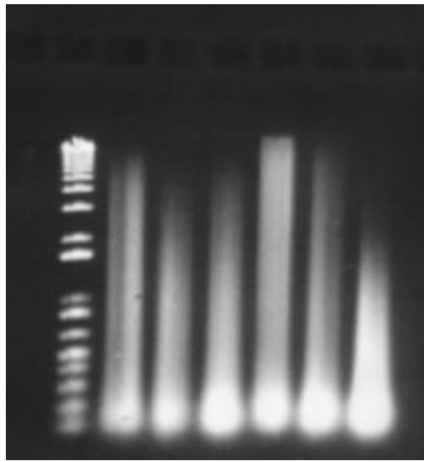
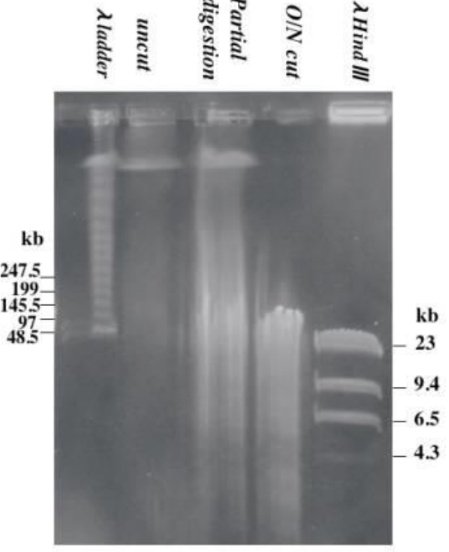
What could have happened to this gel (top)
* If there is a DNA smear at the top of the gel, then the enzyme only possessed partial activity. You must check the reaction conditions because there may be inhibitors present.
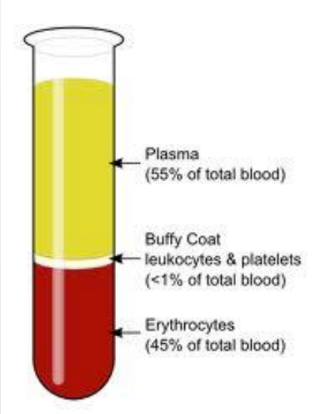
What are the parts of a blood specimen?
Plasma = may contain some genetic material (used for HIV)
Buffy coat = DNA (WBCs &platelets)
Eryhtrocytes = RBCs
What are the steps of an organic DNA isolation? (w/purpose of each reagent)
Lyse the cell (using detergents/proteases) (break the cell contents open)
Acidification if needed (via acetic acid) (if pH needs to be lowered)
Mix lysate with PCI reagent
Forming upper aqueous (DNA) & lower organic phase
Separate aqueous phase
Add ammonium acetate or sodium acetate to encourage precipitation (due to salts)
Add 100% ethanol (promotes DNA precipitation because it’s insoluble in alcohol)
Incubate at –20 to –70 (freezer) (further encourages DNA precipitation)
Centrifuge & pour out supernatant
Wash DNA pellet with 70% ethanol (dissolves salt and not DNA)
Resuspend DNA in TE buffer or water (DNA dissolved and ready for use)
What is the purpose and workflow of ethanol precipitation of DNA
A technique for purifying and concentrating DNA from an aqueous solution
The 100% ethanol promotes DNA precipitation because it is insoluble in alcohol
The 70% ethanol dissolves the salts ONLY without dissolving the DNA
* You first want to encourage the most DNA precipitation as possible, once this is achieved, the salt is removed so the DNA alone can be extracted
Add Salt (sodium acetate) to neutralize DNA’s negative charge
Add cold 100% ethanol (precipitates DNA out of solution & cold enhances it)
Incubate at -20C (allow precipitation to complete)
centrifuge to form pellet of DNA
Wash with 70% Ethanol (dissolves salt and not DNA)
centrifuge again to repellet DNA
Air dry pellet to remove ethanol
resuspend DNA in TE buffer or nuclease-free water
What are the steps of solid phase DNA isolation? Purpose of each reagent
Qiagen
1. Lysis using AL (L = lysis) buffer and proteinase K
Disrupting cells open & stops proteins that can degrade the DNA
2. Incubation at 56 degrees
Accelerates protein breakdown (Proteinase K digests better)
3. Addition of 100% ethanol
Encourages DNA precipitation
4. Addition of AW1/AW2 (W = wash) buffers
First wash removes proteins/contaminants
2nd wash removes the salt/contaminants
5. Elution with AE (E = elution) buffer
DNA released from silica membrane (DNA released for use)
Compare/contrast the spin-column method to the magnetic bead (Chelex) DNA isolation method
Chelex reagent/resin = Used in DNA purification where Chelex beads bind to the cellular debris after cell lysis. Allowing the DNA to be in the supernatant (used in forensics)
Spin-column =
Magnetic (Chelex) | Both | Spin |
|
|
|
What is Salting out
Salting out = inorganic DNA extraction = purification of nucleic acid by precipitating proteins and other contaminants with high salt at low pH
An alternative to using Phenol (toxic reagents)
Low-pH & high salt conc. Causes proteins to be precipitated and DNA left in solution.
DNA is separated and then precipitated in isopropanol (ultimately resuspended in TE buffer/water)
What are the steps to a DNA isolation using a Qiagen spin column
Lyse cells (detergent protease)
Add to column, spin (DNA binds to matrix & waste flows through)
Wash, spin (removes contaminants from column)
Add elution buffer and spin (low salt will release the DNA from the column into a new clean tube)
How are gel-based methods used to determine quality/quantity of DNA preparations?
Quantity = intensity of gel bands
Via densitometry
Quality = no smearing on gel & high molecular weight bands
Excessive smearing means there is degraded DNA
How are spectrophotometric methods used to determine quality/quantity of DNA preparations?
Spectrophotometric = instrument used to measure the absorbance of light at a particular wavelength (QUALITY + QUANTITY)
Nucleic acids absorb light at 260nm
Proteins absorb light at 280nm
Expect a purified sample to have a high A260 and low A280
260/280 ratio indicates QUALITY
Low = protein contamination
High = other contamination
Nanodrops give you the DNA concentration which gives you QUANTITY
Can't distinguish between DNA & RNA
How are fluorometric methods used to determine quality/quantity of DNA preparations?
Fluorometric = Binding fluorescent dyes to DNA and detecting it via a fluorometer
More sensitive than spectrophotometric method (QUANTITY)
Can distinguish between DNA/RNA/contaminants
Good for very SMALL amounts of DNA (smaller than nanodrop)
Not affected by phenol, EDTA, protein, and high salt contamination
How do you calculate concentration and yield of DNA from a preparation
Concentration = amount/volume
Yield = (starting DNA/RNA concentration) / (ending DNA/RNA concentration)
How do you read a spectrophotometric curve (what does it mean)?
A high 280 wavelength means that there is a high amount of purity in the sample
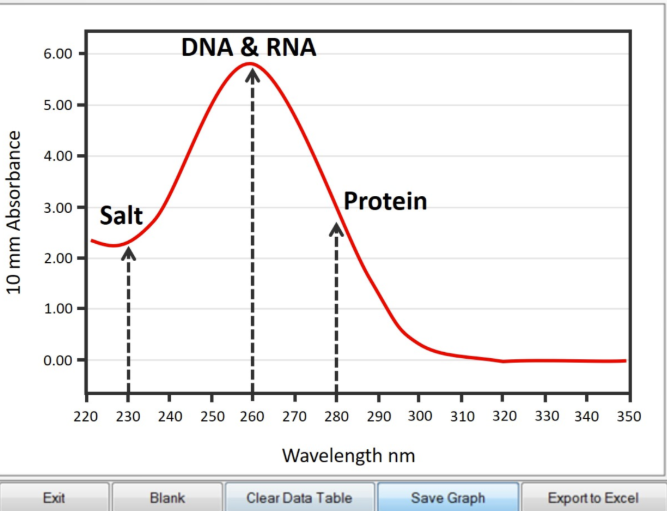
How does a nanodrop give you DNA’s concentration/purity?
Concentration = ng/ul
Purity = A260/280
What changes can you do when PCR has no target bands
For little to no target band:
1. Check your math
2. Verify all components are present
3. Verify primer design
4. Consider if DNA sample has inhibitors (amplified successfully in other reactions?)
5. Lower annealing temperatures
6. Add more template or cycles
7. Add more magnesium
8. Use PCR enhancers or higher denaturation temps for GC-rich targets
What can you do if your PCR has non-specific products
For non-specific products:
1. Increase annealing temperature
2. Reduce primer concentrations (especially if there are abundant primer dimer complexes)
3. Shorten annealing and extension times
4. Reduce cycles or template
5. Reduce magnesium
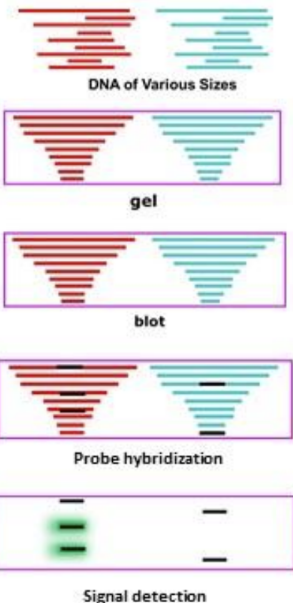
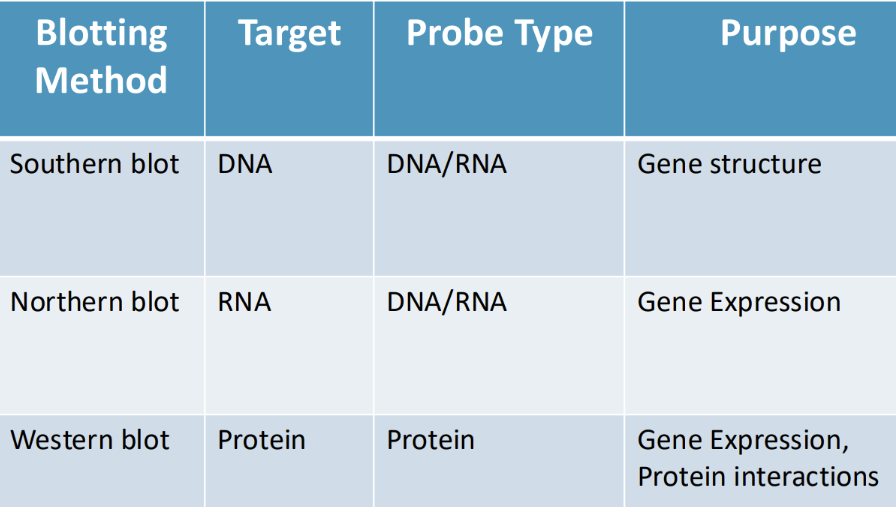
What are the steps to southern blotting
Southern blot steps:
Restriction enzyme digest
DNA is isolated and then cut
Electrophoresis of agarose gel
Fragments are separated by gel electrophoresis, denatured in the gel
Transfer DNA to nitrocellulose membrane (blotting)
Fragments are transferred to a solid support membrane
Hybridize probe to plot
DNA fragments on the membrane are exposed to a labeled probe that is complementary to the region of interest
Probes are usually larger (allow to see individual bands)
Wash blot
Detection of probe signal
The signal of the probe is detected to indicate the presence or absence of the sequence of interest
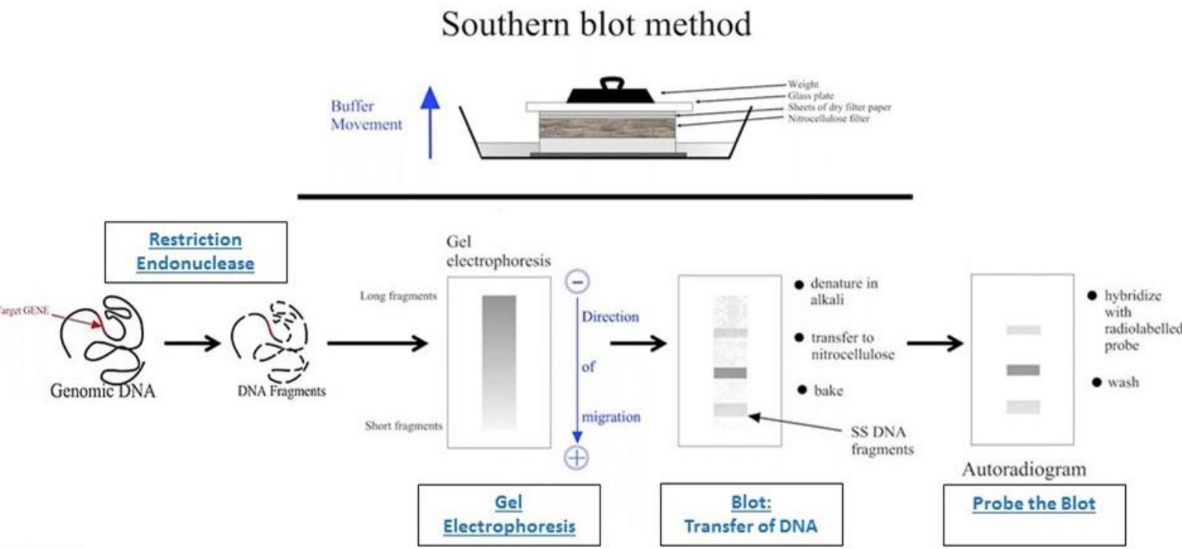
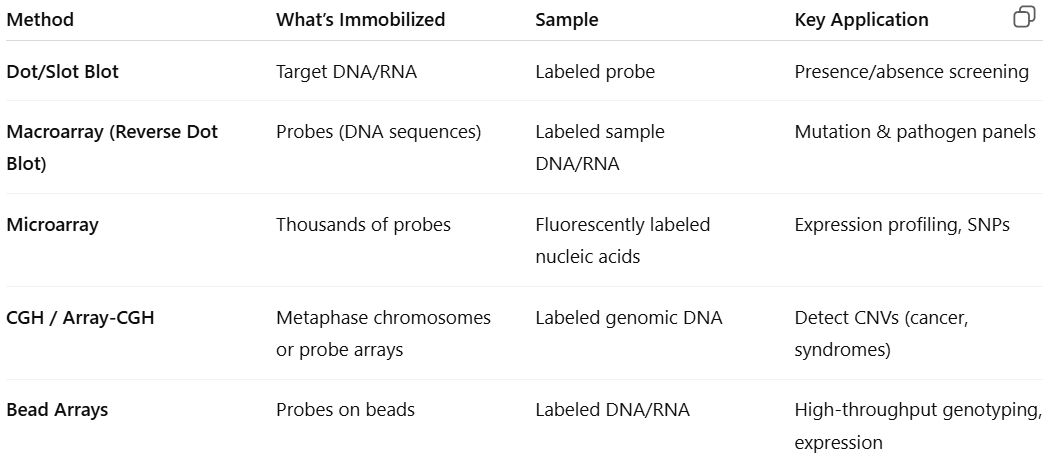
What is dot/slot blots
Dot/slot blots: target DNA/RNA is deposited directly on the membrane by means of various devices (ex: vacuum system)
Applied to expression, mutation, and amplification/deletion analyses
Determination of size is not required
Most efficient on less complex samples
Dot blots = target is deposited in a circle or dot
More useful for multiple qualitative analyses where many targets are being compared (mutational screening)
Ability to test and analyze larger numbers of samples at the same time
Slot blots = target is deposited in an oblong bar
More accurate for quantification by densitometry scanning because they eliminate the error that may arise from scanning through a circular target
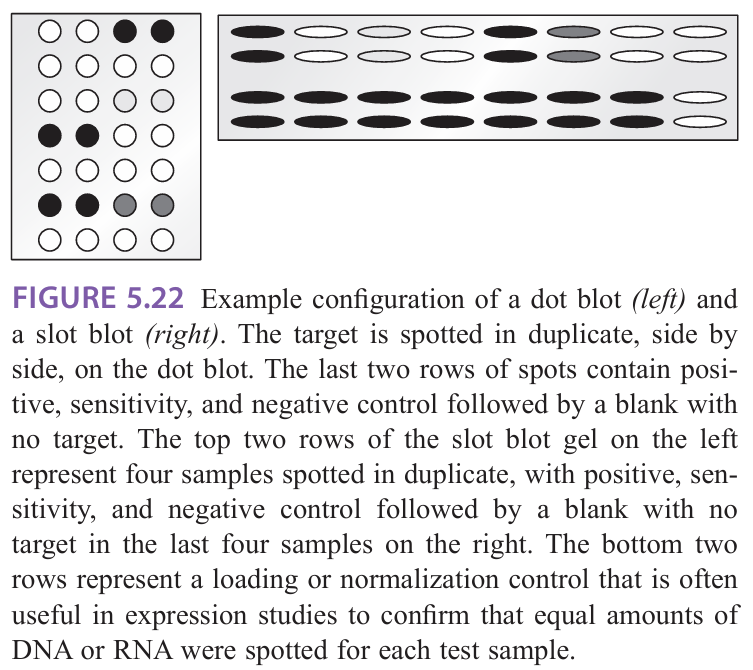
What is macroarrays (reverse dot blot)
Macroarrays (reverse dot blot): many different unlabeled probes are immobilized on the membrane, and the test sample is labeled for hybridization with the immobilized probes
a known sequence is immobilized at a known location on the blot and the amount of sample that hybridizes to it is determined by the signal from the labeled sample
Limited by the area of the membrane and the specimen requirements
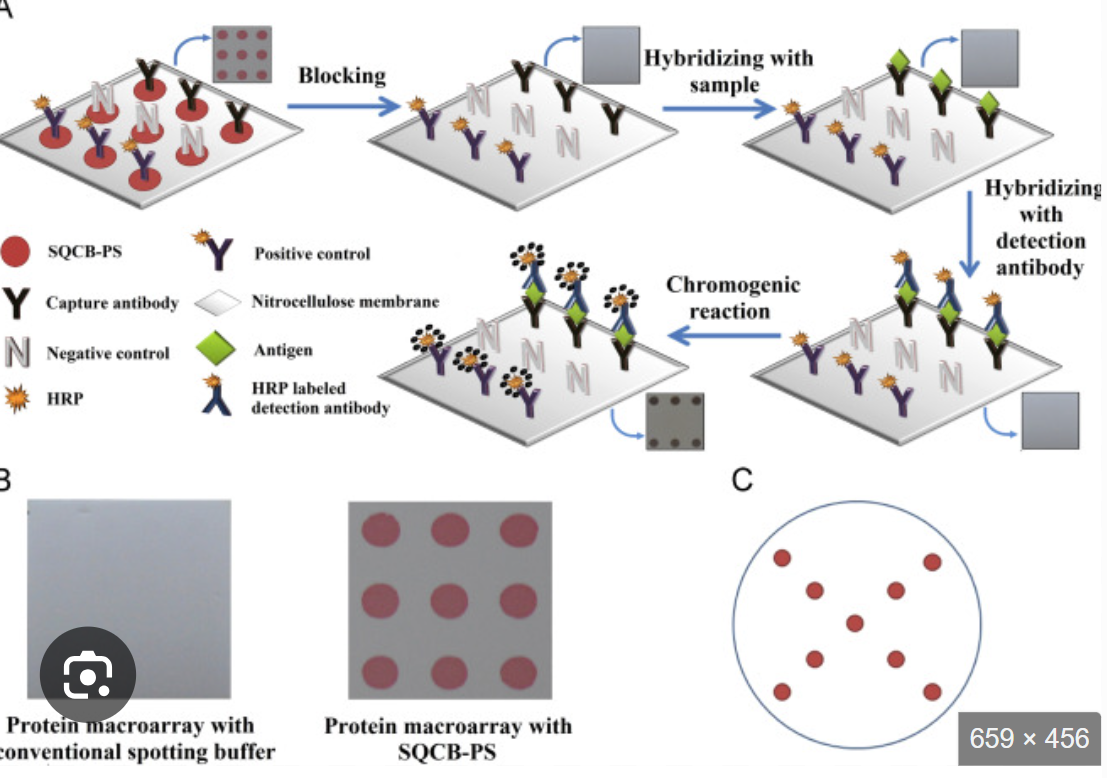
What is microarrays
Microarrays:
Tens of thousands of targets could be screened simultaneously in a very small area by miniaturizing the deposition of droplets
Array targets immobilized on glass slide
Targets can be DNA, RNA, or protein
Requires fluorescent reader and analysis software
Probes are immobilized on a solid support
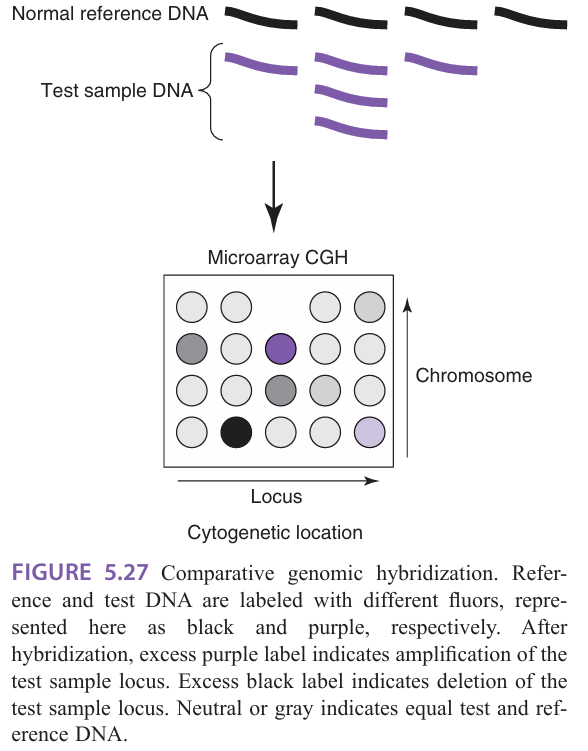
What is comparative genome hybridization (CGH)
Comparative genome hybridization (CGH): designed to test DNA
Used to screen the genome or specific genomic loci for deletions and amplifications
Genomic DNA is isolated, fragmented, and labeled for hybridization on the chip
Provides higher resolution and more defined genetic information than traditional cytogenetic analysis
Limited to the analysis of loci represented on the array
Advantage = can be performed on fixed tissue and limiting samples
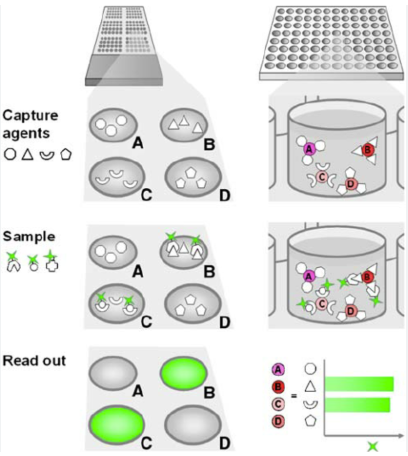
What is bead arrays
Bead arrays: immobilize probes with beads, allowing hybridization of the targets in the bead suspension
Used for protein and nucleic acid targets
Available for infectious disease and tissue typing
Beads are color coded with a particular shade of red fluorescent dye so you can distinguish specific probes carried on different beads
What are the concepts and applications of the following hybridization method: Dot/slot, macroarrays, microarrays, CGH, and bead arrays
look at pic
What are applications of southern blots
Applications of southern blots: (southern blotting is “old school”)
Genetics, oncology (translocations, gene rearrangements)
Detection of repeat expansions (FXS, Huntington)
Typing/classification of organisms
Cloning/verification of cloned DNA
Forensic, parentage testing (RFLP, VNTR)
What is the purpose and probe type of Southern, Northern, and Western blots
picturezzzzzz
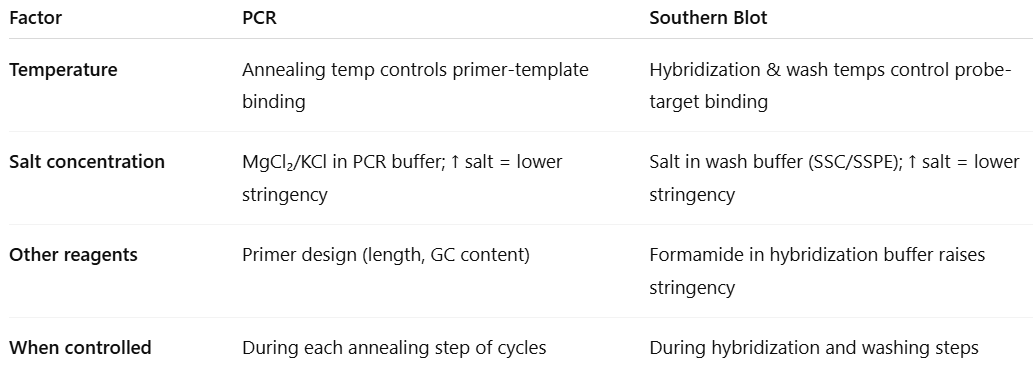
What are factors that affect stringency in blotting
Factors that affect stringency:
Temperature of hybridization
High temperatures = higher stringency
Low temperatures = lower stringency
Wash temperature
high temperatures = higher stringency
low temperatures = lower stringency
Hybridization time
more time = lower stringency
less time = higher stringency
Wash time
lower time = lower stringency
higher time = higher stringency
Salt concentration of hybridization buffer
Keep hybridization solution low
High salt = lower stringency
Lower salt = higher stringency
Concentration of denaturant (formamide) in the buffer
Formamide lowers the optimal hybridization temperature
More formamide = more stringency
Length and nature of probe
Long probe or high GC bases = binds in more stringent conditions
Require longer hybridization times
Short probe or high AT bases = binds in lower stringent conditions
Require lower hybridization times
Increased probe concentration = increased sensitivity of analysis
Ideal conditions = calculated with Tm of probe sequence (and Cot)
How does stringency relate to probe binding
High stringency = more demanding of probe/target complementarity and length
If too high, the probe will not bind to target
Low stringency = more forgiving binding
If too low, the probe will bind to unrelated targets
How is stringency in PCR and southern blotting similar
look at the pic
How should specimens be handled and processed?
Specimen handling:
Preanalytical error can occur if specimen handling, storage, and processing is not done properly
Each assay must have a list of acceptable specimen types (must be validated)
Each specimen must meet established criteria
Method of collection, type, storage conditions, age
Labeling is CRITICAL (at least two identifiers must be present
Name, DOB, patient ID#, specimen ID or requisition #
Processing specimens:
Label (label on tube must contain some of the info. Below)
Name, DOB, age, sec, lab ID, accession #, doc name, collection date with collector initials, test requested, type/source of specimen,
Match = specimen label with paperwork label
Compare = test ordered and specimen type
Can this specimen be used for requested test
Check = shipping and storage (was it done correctly?)
What are some notes for when analyzing specimens
Specimen:
Specimens of minimal cellular content are often analyzed
Cross-contamination MUST be avoided
Specimen is inspected for hemolysis
Lysis of RBCs releases hemoglobin (PCR inhibitor)
If WBCs lysis happened, DNA/RNA yield is reduced
Solid tissues are best analyzed from fresh or frozen tissues
Quality of nucleic acid from fixed tissue depends on the fixing process and the fixative used
What are safety precautions for handling specimen samples? 2 types?
Safety precaution:
All specimens are potentially infectious and should be handled as if they were (standard precautions)
Use PPE
Transmission-based precautions = respirators for airborne or contact transmissible agents
Contact precautions = designed for direct patient care
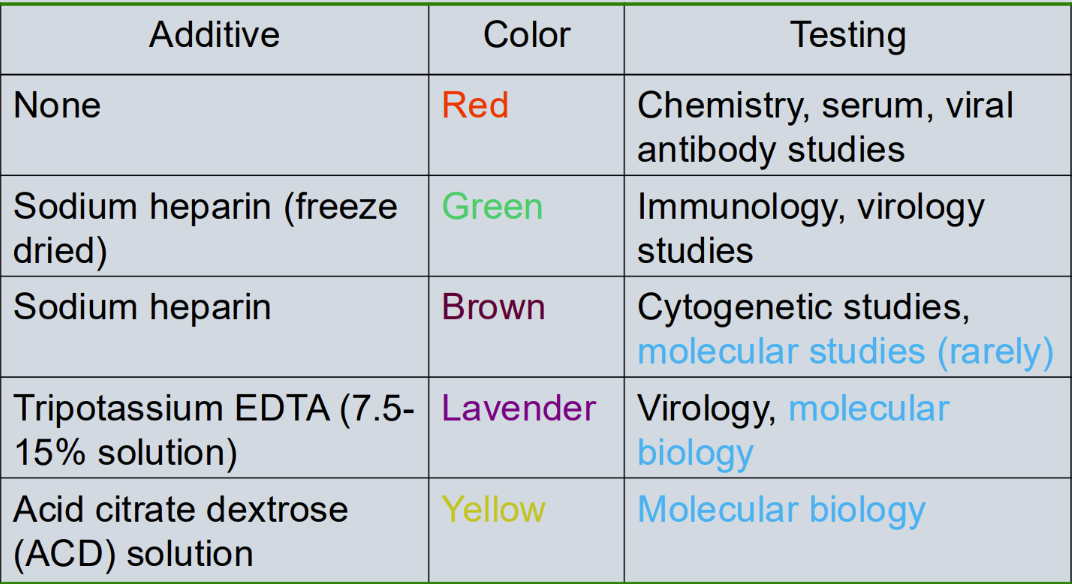
What are the collection tubes used in molecular testing
Tripotassium EDTA (7.5-15% solution) = Lavendar (MOST COMMON)
Testing = virology and molecular biology (Plasma)
Acid citrate dextrose (ACD) solution = yellow
Testing = molecular biology
Sodium heparin = Brown
Testing = cytogenetic studies and molecular studies (rarely)
Specialized tubes:
Plasma Prep Tubes (PPT) = molecular viral testing
PAXgene blood RNA tube = RNA isolation
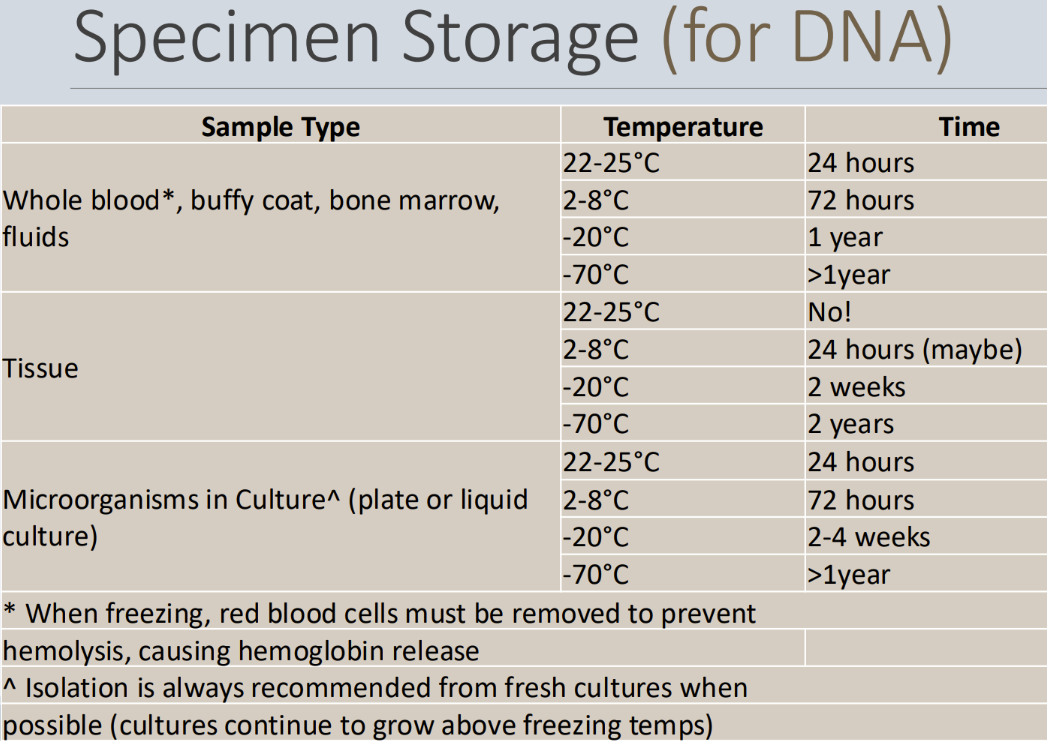
How should specimens be stored if they’ll be used for DNA
Specimen storage that will be used for DNA...
Isolation is ALWAYS better from fresh cultures when possible because DNA has a longer longevity
Can't freeze whole blood, if you do, you need to remove RBCs first
because the hemoglobin is a PCR inhibitor
What is dried blood spots? storage processing?
Dried Blood Spots (Guthrie Cards/FTA cards) VERY STABLE BLOOD STORAGE
Dried blood on thick filter paper
Can be used for isolation of DNA in newborn screenings (for future genetic testing)
Processing = spots must be thoroughly dried
Removal of blood done with saline (20ul of blood is good)
Shipping = keep ambient and dry (use sealed bag or envelope)
Storage = stable indefinitely at room temperature
How should specimens be stored if they’ll be used for RNA
Specimen storage that will be used for RNA...
Goal = to keep as cold as possible
Limited temperature range because RNA is labile
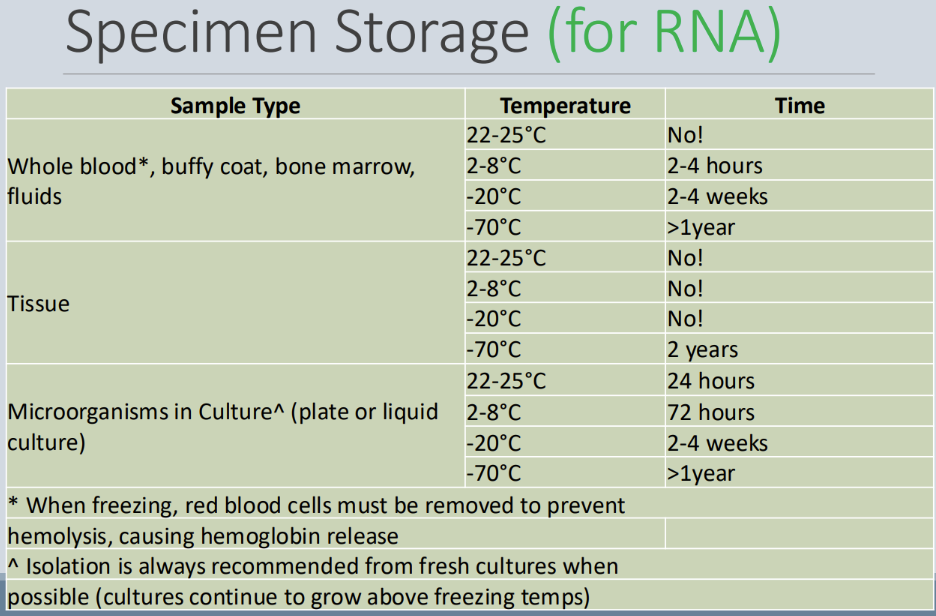
How should specimens be stored if they’ll be used for nucleic acid
Nucleic acid storage...
After specimen has been processed
DNA can be stored for longer than DNA
Freeze dried and collection paper dried DNA is stable at room temperature
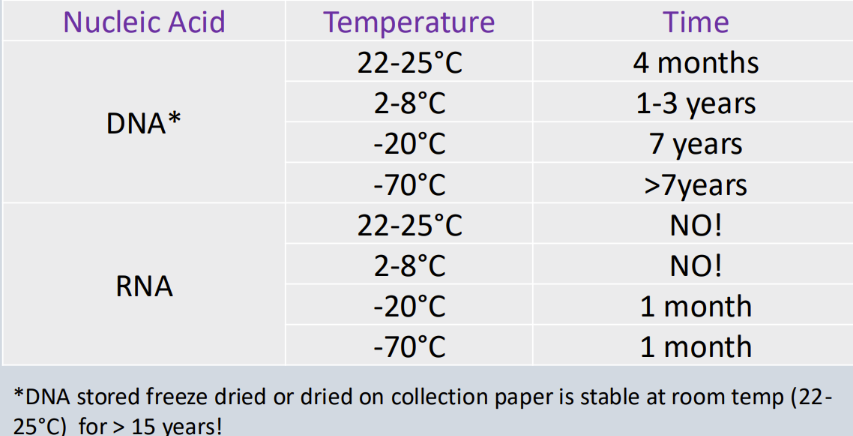
What are the requirements of quality assurances
Quality assurance: lab operations
Most molecular labs seek accreditation by the College of American Pathologists (CAP)
Clinical laboratories are regulated by the government via the CLIA
Defines quality standards
All aspects of pre-analytical, analytical, and post-analytical processes must be monitored in the QA program
Not government run but can be stricter than the gov.
Labs must have a Quality Assurance program
All clinical tests must have Quality Control (QC) (specific to a particular test/run)
Quality monitoring of a specific assay
QC sample has characteristics very similar to patient samples
Has known value
Is treated and tested in the same manner as patient samples
What are the requirements for temperature checks
Temperature checks
Refrigerators and freezers used to store patient material or reagents must be monitored at least daily
Minimum/maximum thermometers can measure the lowest and highest temperatures between monitoring points
Automatic temperature monitoring systems are common, can notify user of out-of-range temps
Heat blocks, incubators, ovens, water baths must also be monitored daily or upon usage for an assay
All thermometers must be NIST-traceable
Meaning they are validated against a reference thermometer (highest standard)
What is calibration?
Calibration = the adjustment of an instrument or assay result to the actual concentration of a known reference analyte by testing and making the appropriate adjustments
Uses standards (frame of reference) - often in multiple dilutions (standard curve)
The range of the standard curve establishes the reportable range (or AMR)
Any result above/below the AMR must be reported as “greater/less than”
Calibration verification = if calibration is not done on each run, then verification must be done every 6 months or more frequently if major components are changed or reagent lots changed