MIC102 MT2
1/133
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
134 Terms
What is the central dogma of molecular biology?
DNA → RNA → Protein.
Genetic information flows from DNA to RNA (transcription), then from RNA to protein (translation)
What are some of the key enzymes involved in DNA replication? (5)
DNA Polymerase- Adds nucleotides to the growing DNA strand
Helicase: Unwinds the DNA double helix
Primase: Lays down RNA primers for DNA polymerase
Ligase: Seals gaps between Okazaki fragments
Single-strand binding proteins: stabilize unwound DNA
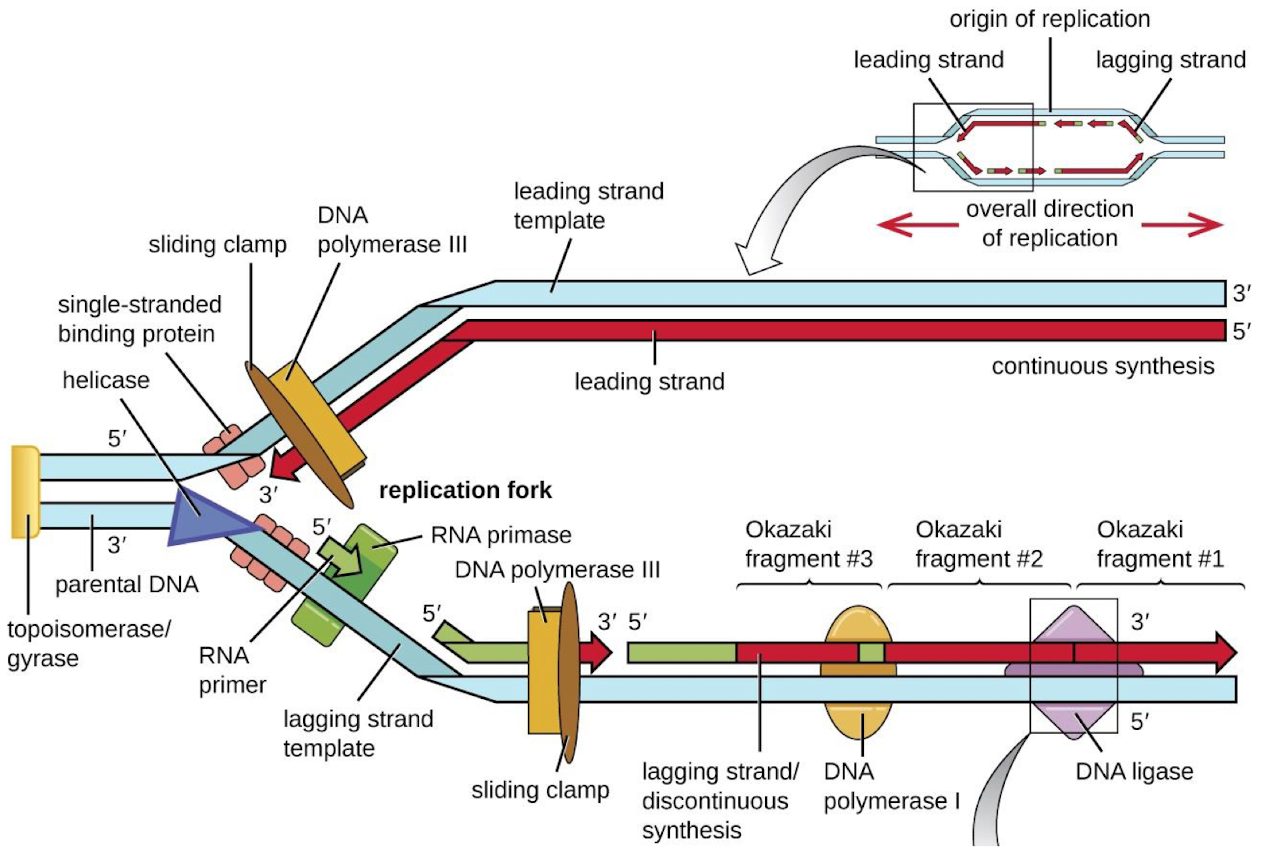
Why does DNA replication require an RNA primer?
DNA polymerase needs a free 3’ hydroxyl group (-OH) to start adding nucleotides
What is the leading vs lagging strand in DNA replication?
Leading strand: Synthesized continuously in the 5’ to 3’ direction
Lagging strand: synthesized in short Okazaki fragments that are later joined by DNA ligase
Where does bacterial DNA replication start and stop?
Starts at the Origin of Replication (OriC): An AT-rich sequence easier to unwind
Stops at the Terminator (Ter): ensures full replication and prevents premature stopping
Why do bacteria replicate DNA faster in rich media?
In nutrient-rich conditions, new rounds of replication begin before the previous one finishes, ensuring rapid cell division
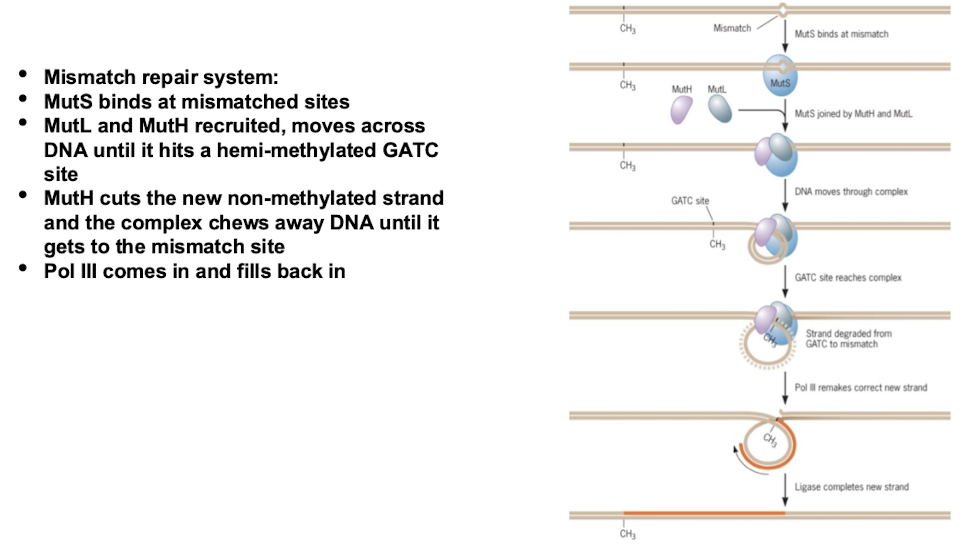
How does DNA mismatch repair work?
MutS protein detects mismatched bases
DNA polymerase fixes mistake
Methylation marks the old (correct) strand as a reference
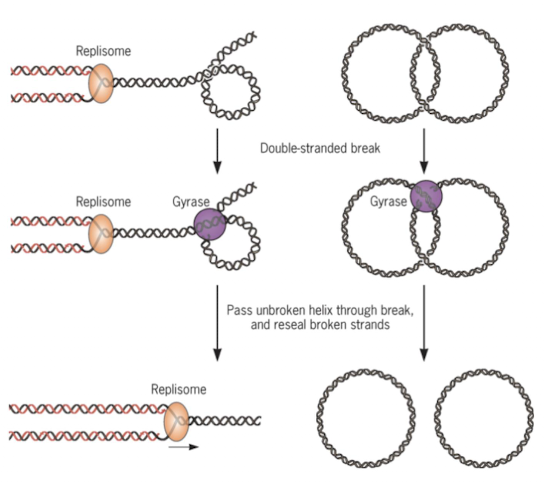
What is the role of DNA gyrase?
Relieves supercoiling tension during DNA replication. It’s a target for antibiotic since humans don’t have DNA gyrase
What does RNA polymerase do?
Synthesizes RNA using DNA as a template (DNA-dependent RNA polymerase). It doesn’t need a primer like DNA polymerase
What are sigma factors and why are they important?
Help RNA polymerase recognize promoters to start transcription. Different sigma factors regulate genes needed under different conditions (e.g. stress response, nitrogen metabolism)
What is the function of ribosomal RNA (rRNA)?
Forms part of the ribosome, which is the site of protein synthesis. It accounts for 90% of cellular RNA
How does translation work?
mRNA provides the code
tRNA delivers amino acids based on codon-anticodon matching
Ribosome catalyzes peptide bond formation to make proteins
What is the function of tRNA?
tRNA reads mRNA codons and delivers the corresponding amino acid to the ribosome
How do bacteria achieve rapid protein synthesis?
Transcription and translation occur simultaneously (coupled transcription-translation), unlike in eukaryotes
What is the difference between factor-independent and factor-dependent transcription termination?
Factor-independent: RnA forms a hairpin loop, causing detachment
Factor-dependent (Rho dependent): The Rho protein releases RNA polymerase from DNA
What is the difference between mRNA, tRNA, and rRNA?
mRNA: Messenger RNA; contains instruction for protein synthesis
tRNA: Transfer RNA; carries amino acids to ribosomes
rRNA: Ribosomal RNA; structural and enzymatic component of ribosomes
What is an operon?
A cluster of genes transcribed together as a single mRNA, allowing coordinated gene expression (e.g., lac operon for lactose metabolism)
What are the 3 ways for gene expression
transcription: controls whether mRNA is made
translation: controls how much protein is made from mRNA
Post-translation: Controls protein activity after it is made
leaky transcription
bacterial genes are never really off, always a little transcription
Constitutive genes (housekeeping genes)
“always” on and required for basic functions
Regulated genes
can be inducible (turned on when needed) or repressible (turned off when unnecessary)
is lac operon regulated or constitutive
regulated but is leaky and always has lactose permease present
lacZ, lacY, LacI functions
lacZ makes beta-galactoside and that makes allolactose
lacY makes lactose permease, which allows lactose to be permeable and move through the membrane
LacI makes a protein/transcription factor that represses the lac operon, thus lac-i down-regulates the lac operon.
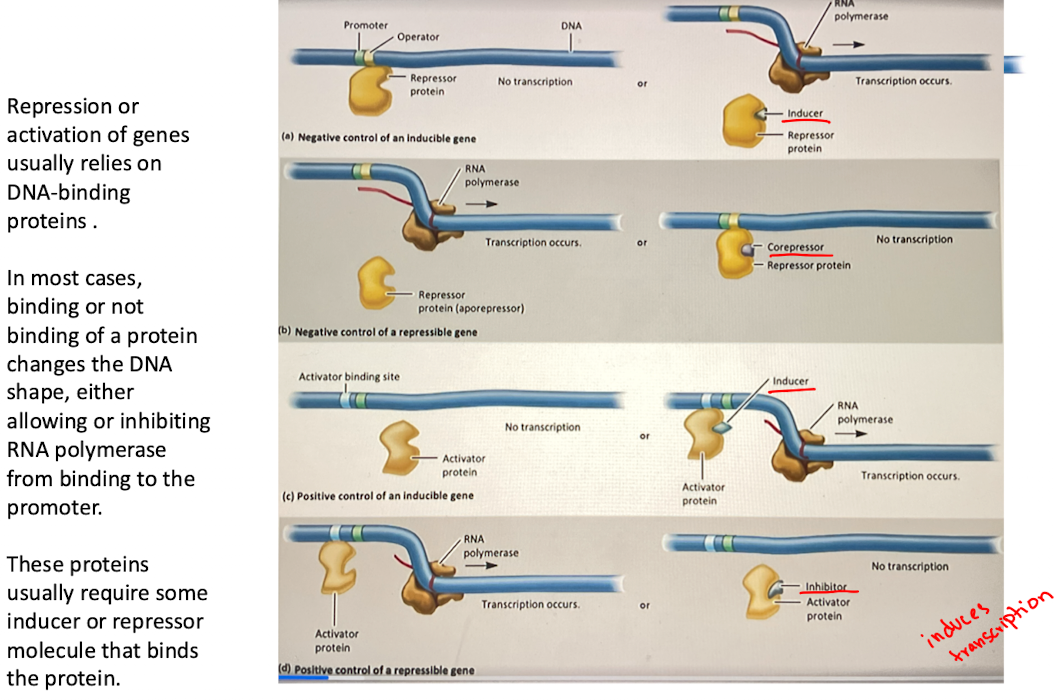
a. Negative control of an inducible gene
A repressor protein binds to the operator, blocking RNA polymerase and preventing transcription
When an inducer binds to the repressor, it changes shape and releases from the operator, allowing transcription to occur
Ex: lac operon
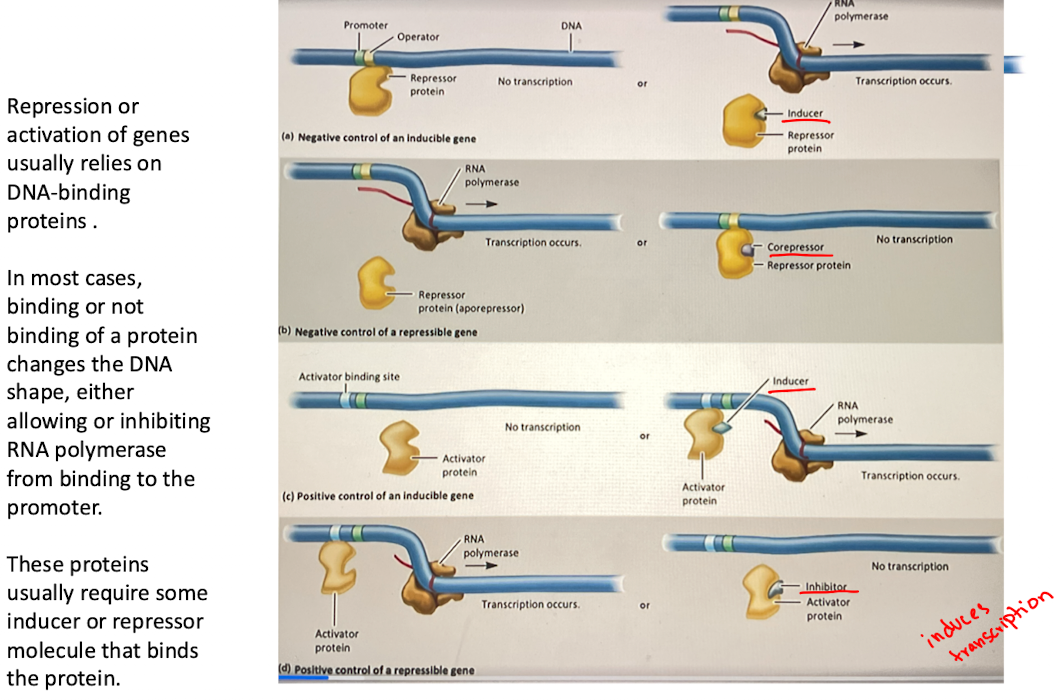
B. Negative control of repressible gene
the repressor protein (aporepressor) is initially inactive, allowing transcription
When a corepresor binds to the repressor, it activates it, and the complex binds to the operator, blocking transcription
ex. the trp operon (trp biosynthesis)
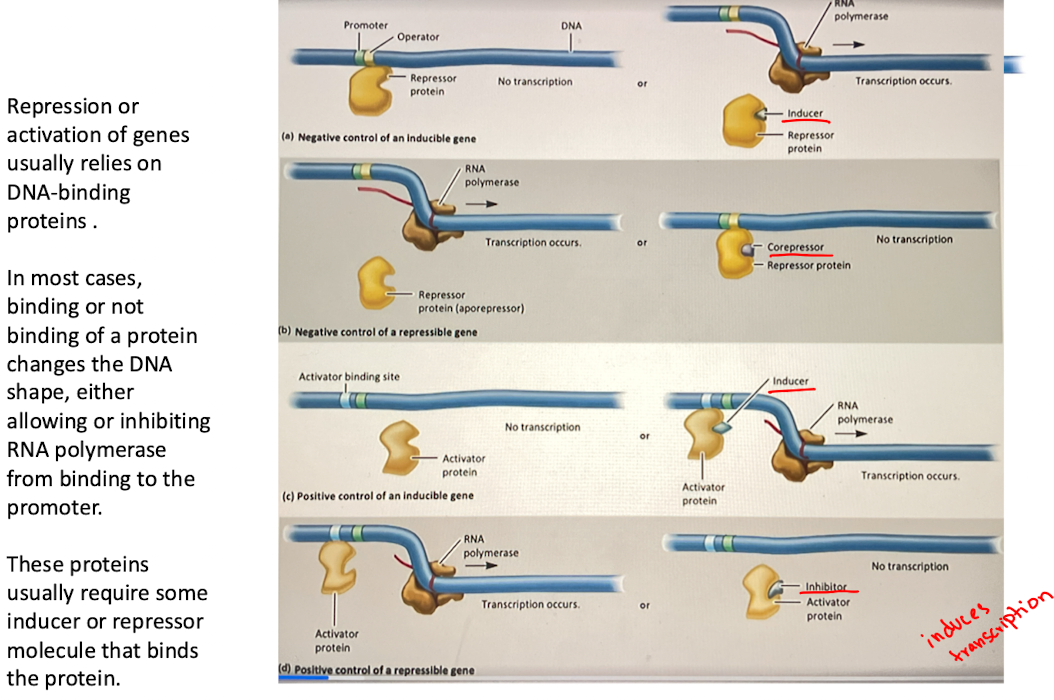
C. Positive control of an inducible gene
activator protein alone cannot bind to the DNA, so transcription does not occur
Inducer binds to activator, changes shape and binds to activator-binding site, recruiting RNA polymerase and inducing transcription
ex. CAP-cAMP system in the lac operon (glucose control)
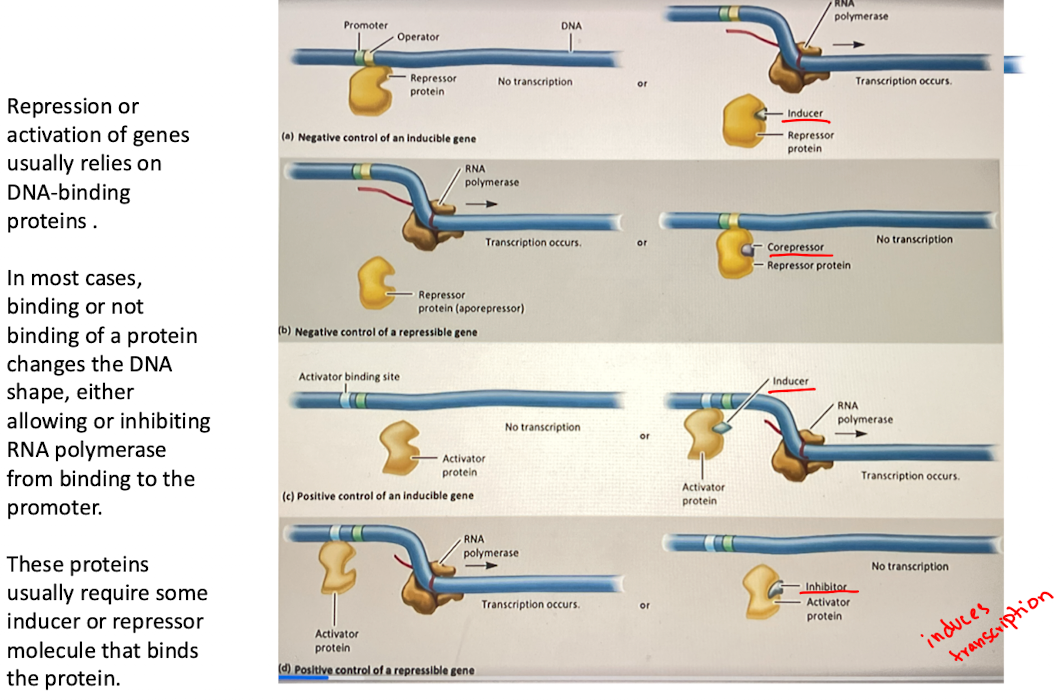
D. Positive control of a repressible gene
activator protein is required for transcription
Inhibitor binds to activator, prevents it from binding to DNA, stopping transcription
ex. certain metabolic pathways where gene expression stops when end product is abundant
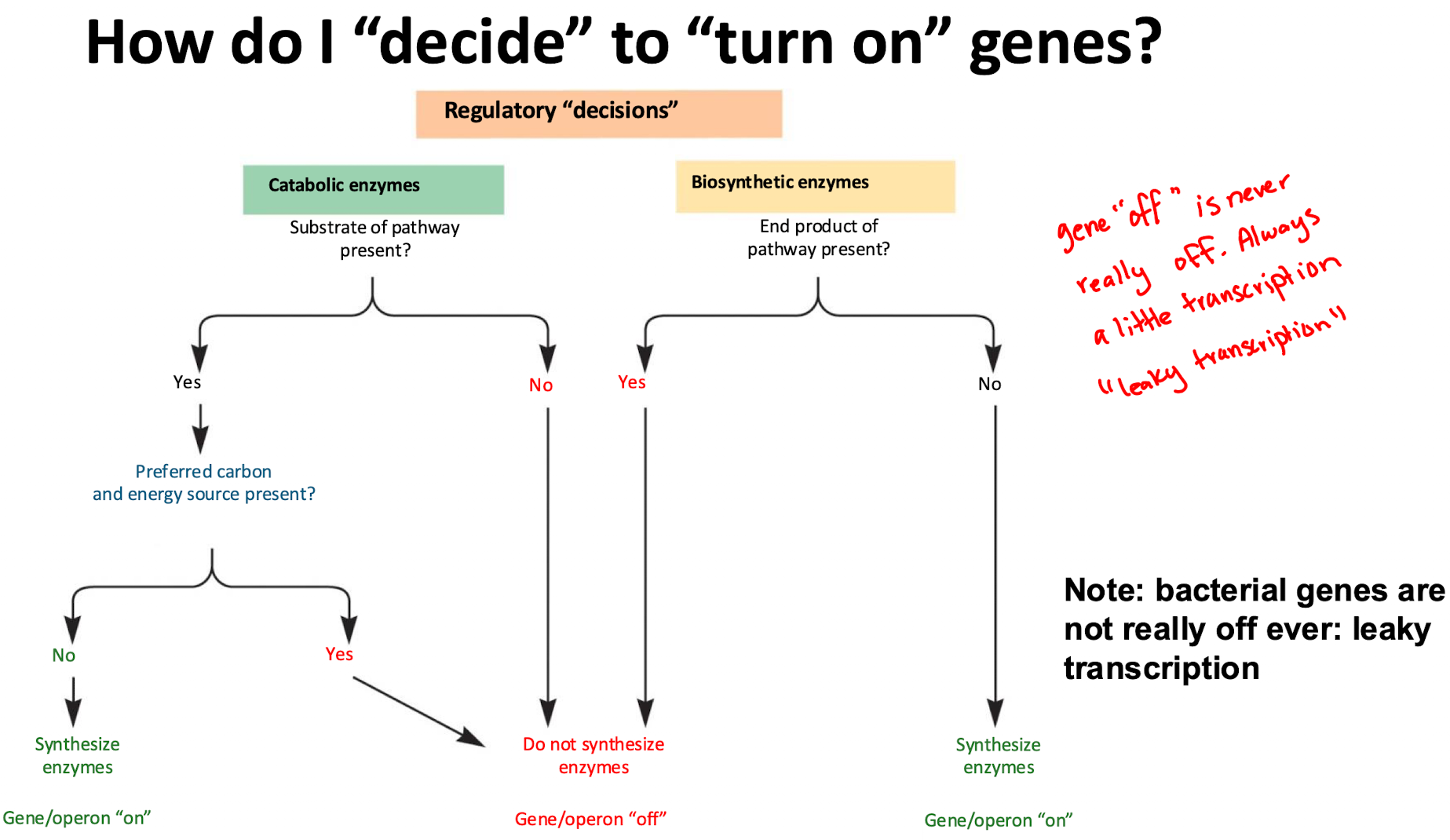
How do bacteria regulate catabolic (breakdown) enzymes?
If substrate (lactose) is present, enzymes are made (on)
If a preferred energy source (glucose) is available, bacteria repress unnecessary enzymes (off)
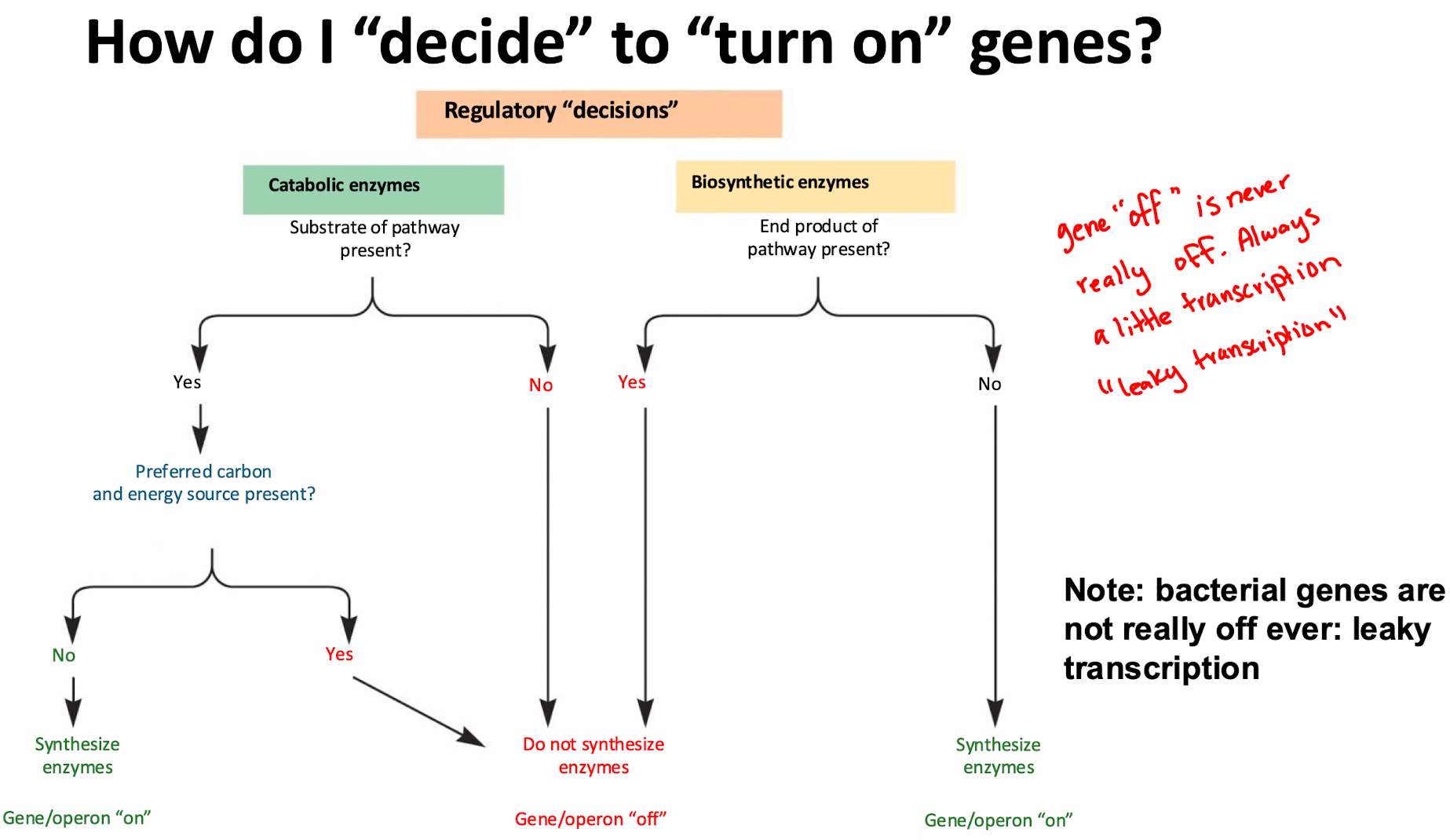
How do bacteria regulate biosynthetic (anabolic) enzymes?
If end product (trp) is available, enzyme production stops (off)
If end product is absent, enzymes are synthesized (on)
How does the lac operon work?
lac operon controls lactose metabolism
Lacl repressor bind the operator and blocks transcription
Allolactose (inducer) binds to Lacl, removing it from the operator, allowing transcription
CAP-cAMP complex enhances transcription but is only active when glucose is low
global regulator in catabolite repression
cyclic AMP receptor protein (CRP or CAP)
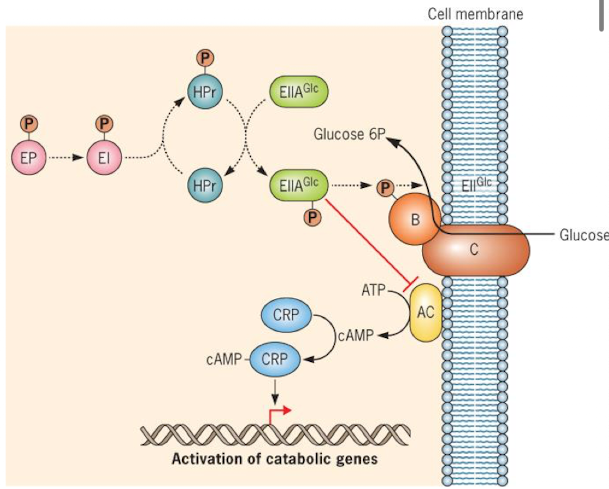
How does catabolite repression regulate the lac operon?
when glucose is present, cell prefers glucose and won’t use lactose
glucose lowers cAMP and adenylate cyclase (AC) activity, so CAP cannot activate the lac operon (no transcription of catabolic genes)
When glucose is low/used up = AC activity resumes, cAMP increases, CAP binds DNA, and the lac operon can be transcribed
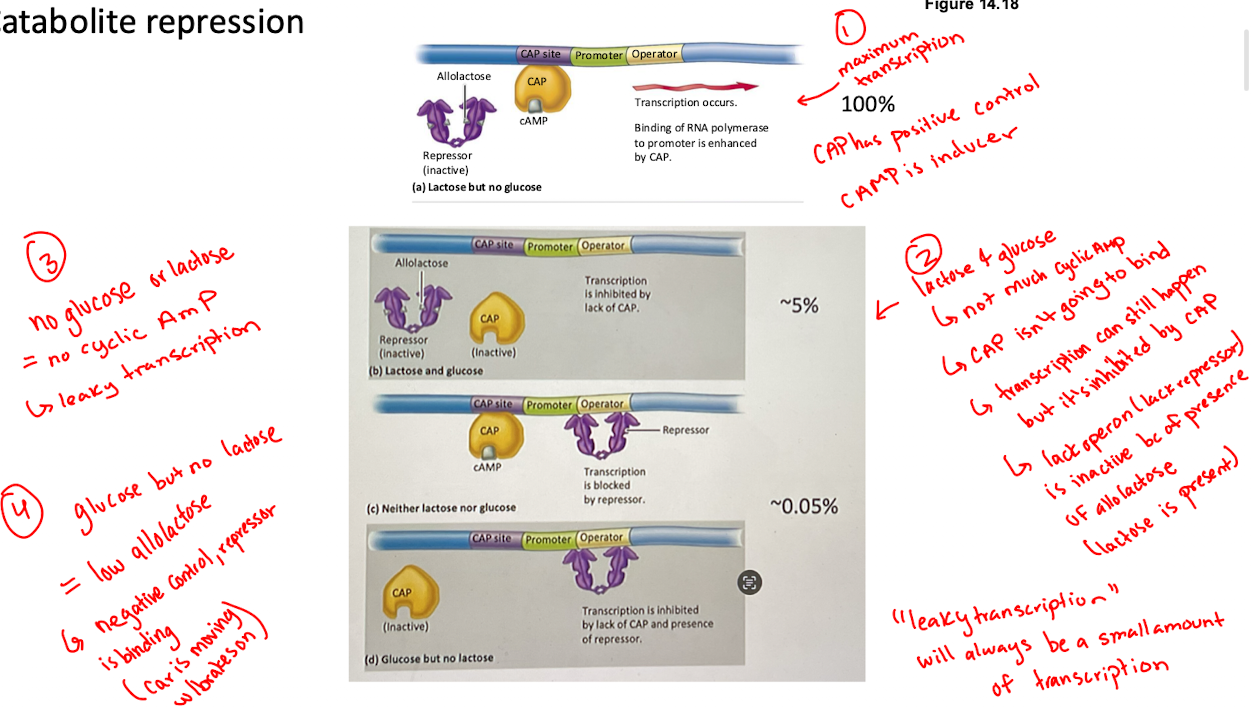
What is attenuation in the trp operon? (if trp is high?low?)
If trp is high, ribosomes move quickly, causing terminator loop in mRNA that stops transcription
If trp is low, ribosome stall, forming an anti-terminator loop, allowing transcription to continue
Attenuation: transcription is terminated shortly after initiation before completing the transcript
Regulatory RNAs (sRNA): Where they bind
6S RNA made in stationary phase binds to sigma-17 RNAP and prevents genes from being transcribed
Some bind DNA directly
Some bind to mRNAs and stop transcription/stability
Some bind to mRNAs and alter translation
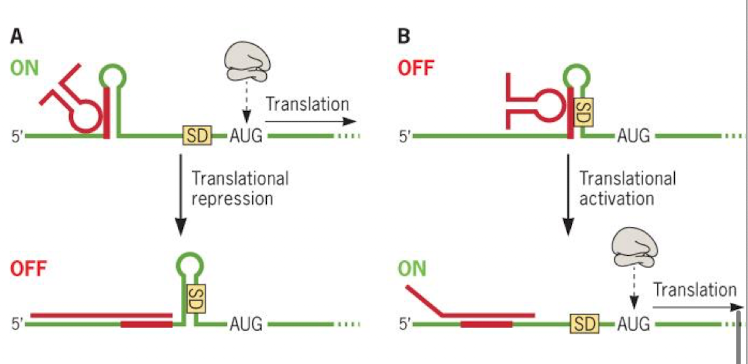
Global regulation: regulon and modulon
Regulon: one regulator affects each operon in regulon; sigma factors control regulons
Modulon: one regulator affects multiple regions; modulo’s control sets of regulons
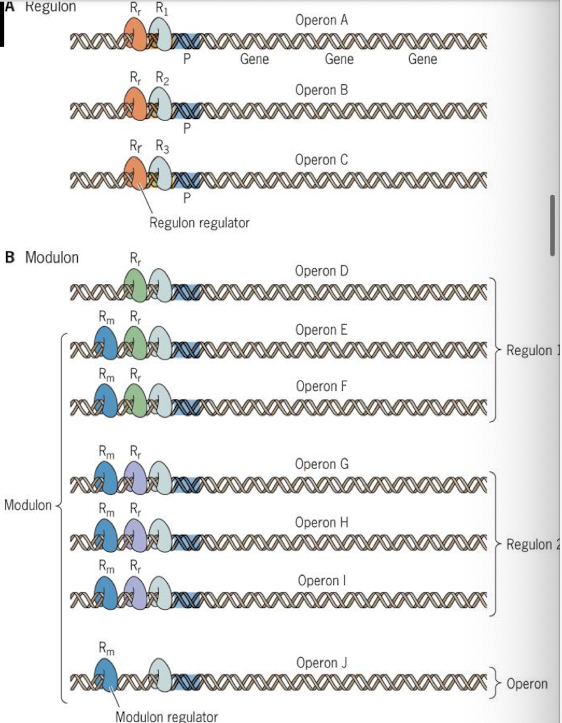
Two component signal transduction systems
Sensor kinase: Histidine kinase + response regulator; transmembrane protein that can sense environmental signals
Response regulator: DNA binding and stress responses; targets gene to be controlled
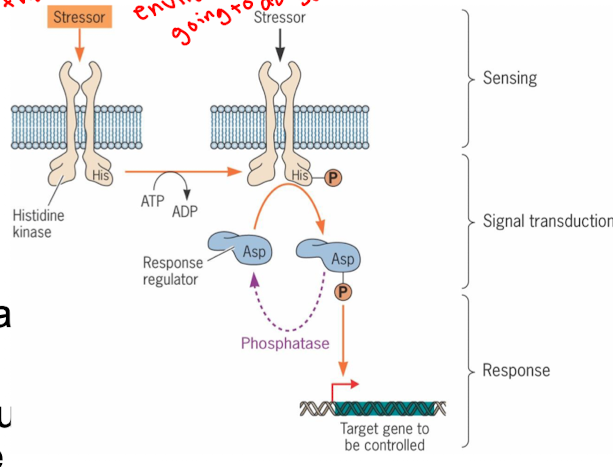
Why is motility highly regulated
cells need to move to gather nutrients or avoid stresses/dangers
flagella rotate counterclockwise to move cells cells better (clockwise rotation causes random tumbling)
chemotaxis
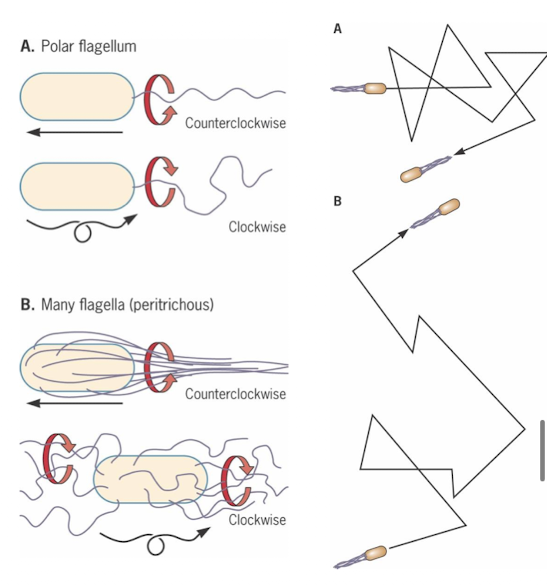
What is chemotaxis and how is it regulated? (CheA, CheY, CheB functions)
Methyl-accepting chemotaxis proteins (MCPs) sense attractants/repellents; bind signal molecules
CheA (sensor kinase): senses MCP conformation
CheY: response realtor, controls flagella rotation direction
CheB: another response regulator, methyl transferase
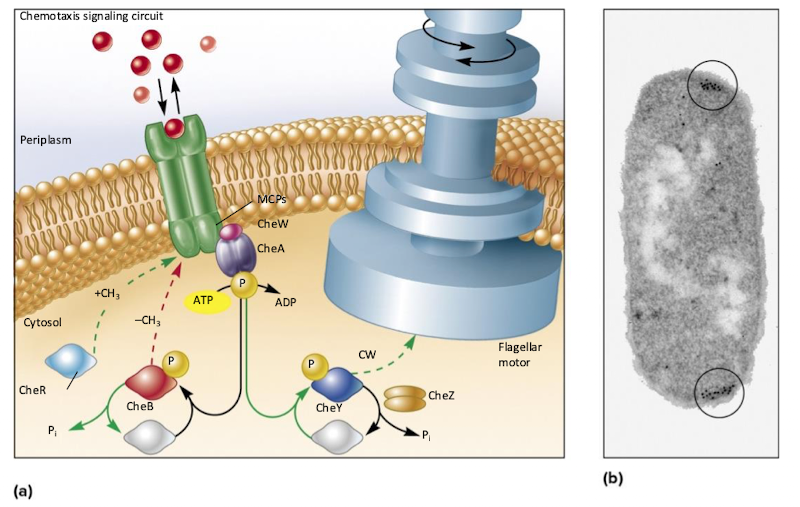
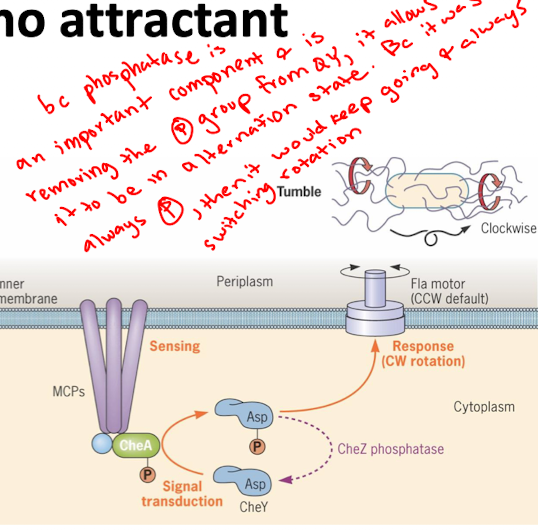
What happens when an attractant is not sensed (CheA, CheY, CheZ)
CheA autophosphorylates and passes the signal to CheY RR
CheY-P makes flagella turn clockwise (tumbles)
CheZ phosphatase dephosphorylates CheY (stops tumbling)
Alternates tumbles and runs
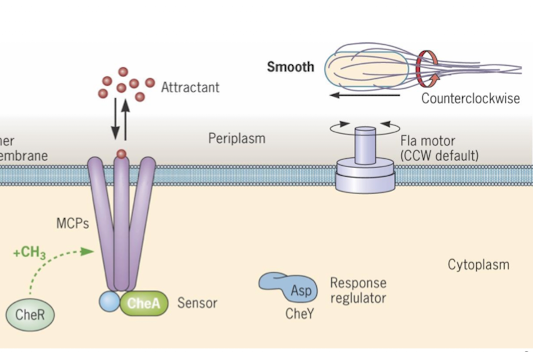
What happens when an attractant (i.e. glucose) is sensed (CheA, CheY, CheZ?)
Attractant binds to MCP, inhibiting CheA autophosphorylation
No active CheA means no active CheY causing counterclockwise flagella motion (run)
Methylations of MCP resets conformation, allowing CheA to activate again, requiring higher attractant levels to inhibit it again
accommodation ensures bacteria moves towards increasing attractant concentration
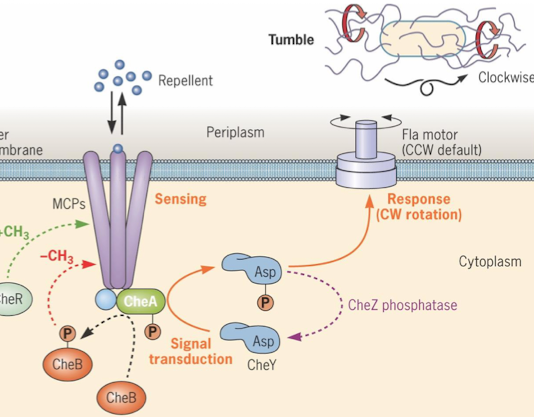
What happens when repellents are sensed?
repellent binds to MCP, stimulating CheA autophosphorylation (tumbles)
high activity Che A phosphorylates CheB, removing methyl groups from MCP (sense smaller quantities of repellent)
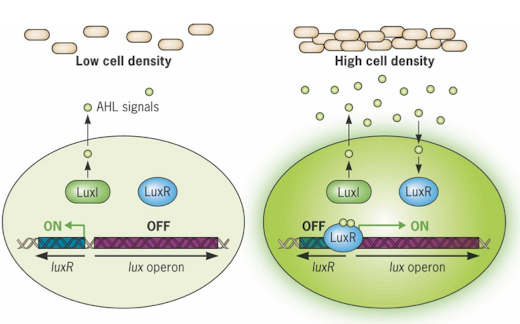
What is quorum sensing, and what is it used for?
Bacteria communicate using autoinducers like homoserine lactones (AHLs) in gram-negatives and specific peptides in gram-positives
Cells wait until population density is high before activating genes
Used for virulence, biofilm formation, light production
How does Luxl-LuxR quorum sensing system work?
Luxl produces AHL autoinders
At high AHL levels, it binds LuxR, activating lux operon
Leads to luciferase production, making bacteria glow
What are the 3 requirements for natural selection?
variation
heritability
Differential reproductive success
Effects of mutations
Neutral: no effect (due to redundancy in genetic code)
Harmful: lose function
Beneficial: Rare but can create new functions that help survival
What are spontaneous vs induced mutations?
Spontaneous mutations happen during DNA replication (error rate is low due to proofreading)
Induced mutations are caused by external factors (mutagens) like chemicals or UV radiation
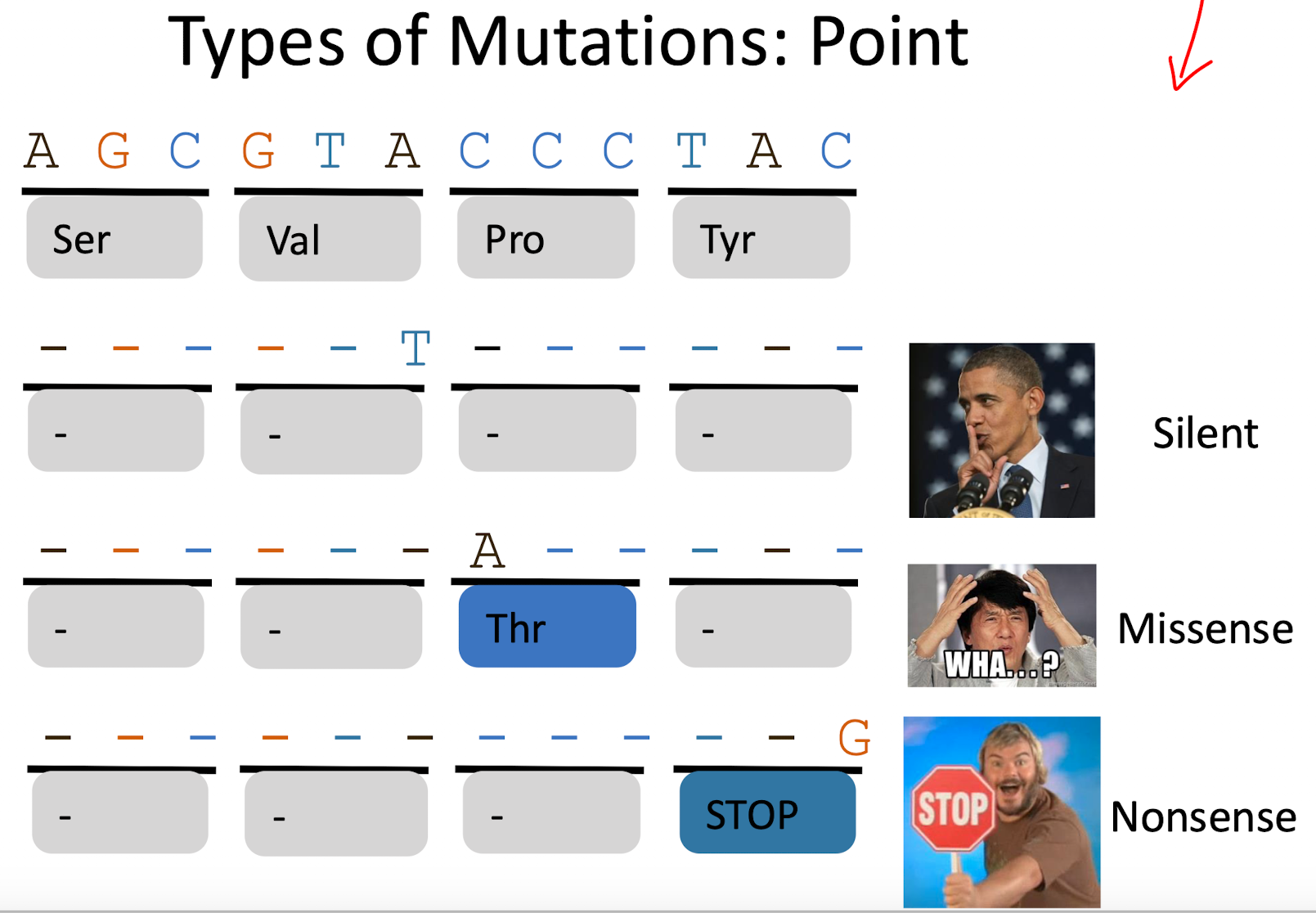
What are point mutations and their effects? (3)
silent mutation: no amino acid change
Missense mutation: one amino acid changes, possible altering function
Nonsense mutation: creates a premature stop codon, stopping protein production
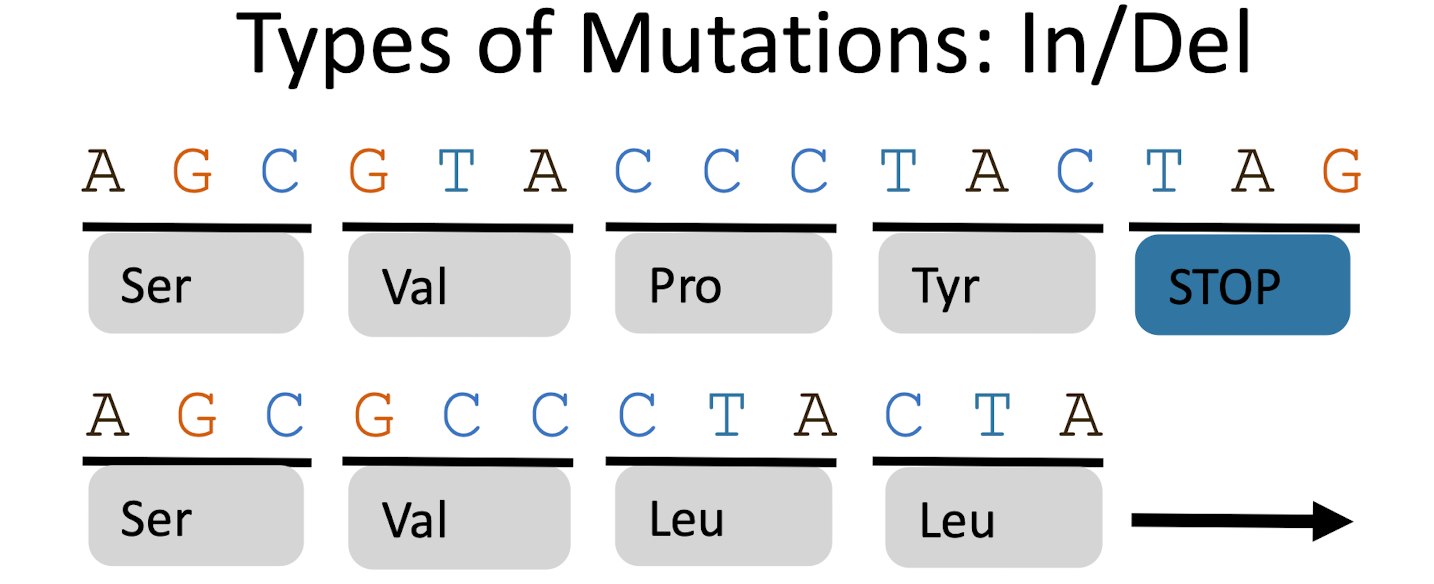
What are frameshift mutations?
Insertions or deletions shift the reading frame of codons, altering every amino acid after the mutation. Often leads to nonfunctional proteins
Mutation definition
any change in DNA sequence inherited by offspring; some have no observable affect on phenotype
Why do bacteria have mutations if DNA polymerase is so accurate?
While error rate is low (1 in a billion base pairs), bacteria reproduce in huge numbers
What are auxotrophs?
Bacteria that lose ability to synthesize a building block or other growth factor.
Are temperature sensitive (TS) mutants and conditional lethal (cannot grow in high temps)
Enrichment (auxotroph screening)
grow in a medium without the component you want to find auxotrophs for
Add penicillin to kill non-auxotrophs
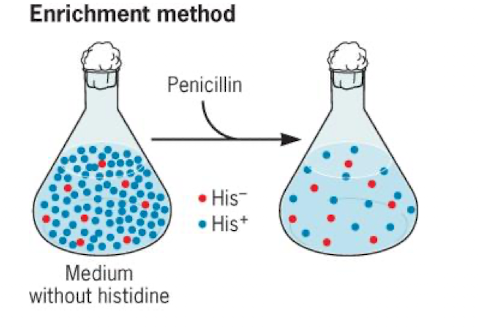
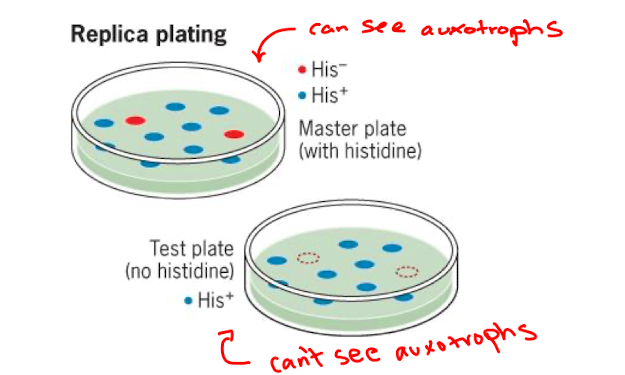
Screening (studying mutants)
plate onto permissive medium
take colonies from master plate and restreak them onto medium without component (colonies that can’t grow on new medium are auxotrophs)
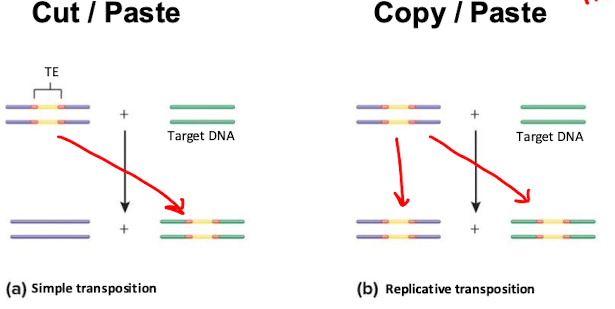
Mobile elements (transposable elements)
pieces of DNA that can move around a chromosome
Non-composite transposon: contain drug resistance gene
Composite transposon: two sets and repeats surrounding other genes
Simple vs Replicative Transposition
Simple: Cut and paste TE into target DNA
Replicative: Copy and paste Te into both target DNA and new cell
What is site-directed mutagenesis and how does it work?
It is a targeted mutation method using PCR with designed primers to introduce specific mutations into a gene, which is then incorporated by homologous recombination
What is complementation and how does it confirm a mutant gene’s effect?
Complementation tests whether a wild-type (WT) gene restores the normal phenotype. If it does, the mutant gene is confirmed to cause the observed effect
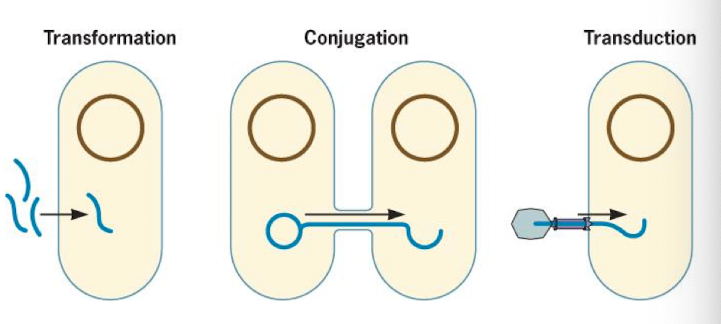
What is horizontal gene transfer (HGT)?
Genetic exchange in prokaryotes that doesn’t rely on reproduction. Small amounts of DNA exchanged
Transformation
Transduction
Conjugation
How is DNA taken up and integrated during natural transformation?
Competence proteins bind dsDNA
one strand is degraded, the other enters the cell
RecA coats ssDNA, finds a homologous sequence, and facilitates recombination into the chromosome
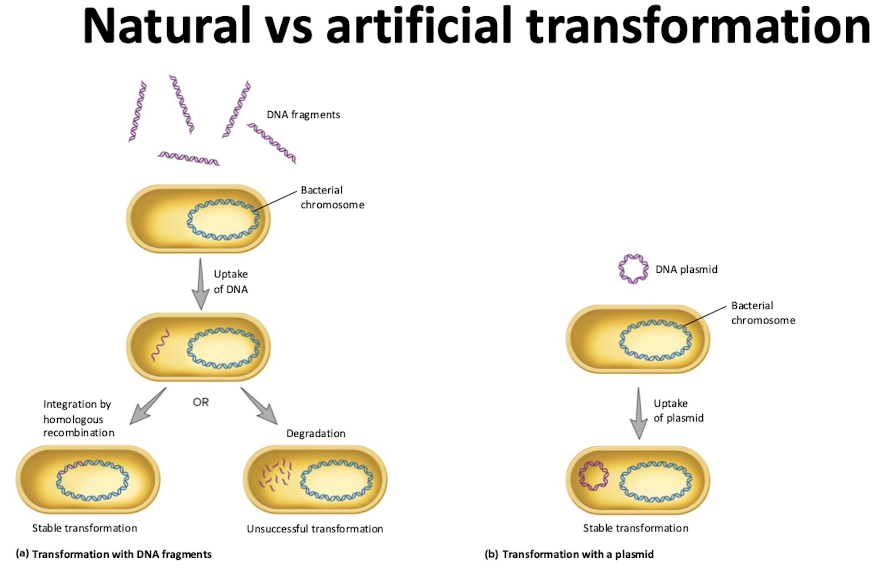
What are some artificial transformation techniques to introduce DNA into the cell?
Heat shock, electroporation, firing DNA coated into a metal into the cell
Typically uses plasmid DNA
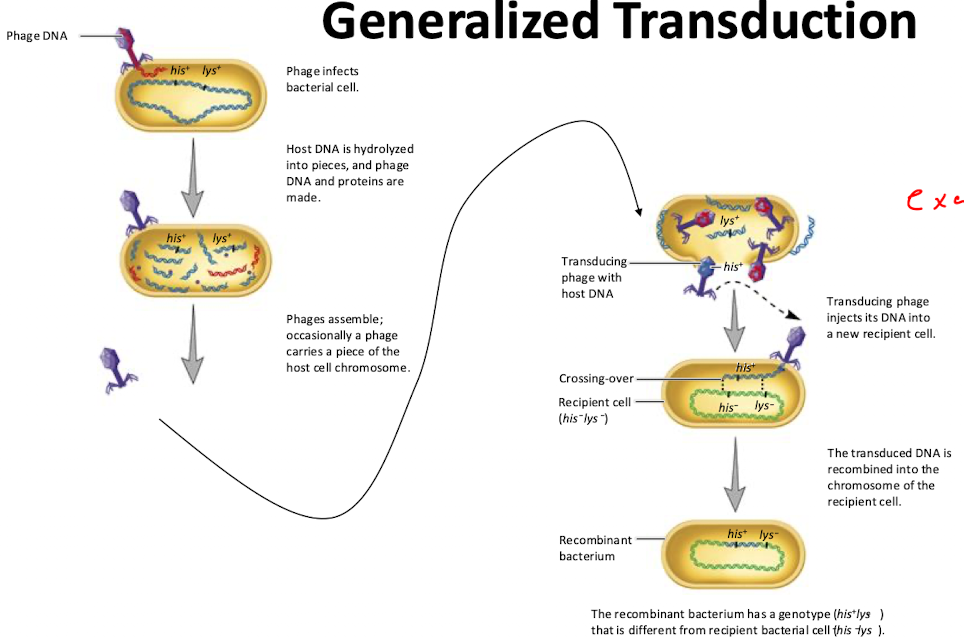
What is transduction?
The transfer of genetic material from one organism to another by a virus.
Generalized Transduction
Process where a phage accidentally packages random bacterial DNA into its capsid instead of viral DNA
During rolling-circle replication, phages use pac sites to package viral genomes but bacterial DNA with similar sites can also be mistakenly packaged (phage P22)
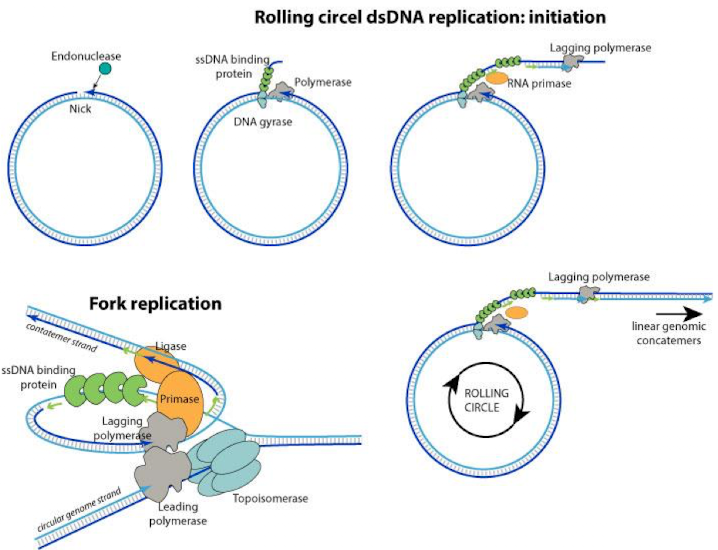
Rolling circle replication
important to phage replication and bacterial conjugation
circular DNA is nicked and unwound allowing polymerase to use exposed 3’ strand to begin synthesis allowing replication to continue around entire molecule if small and minimal supercoiling
What is specialized transduction and which phage is the model for it?
Occurs when a temperate phage (λ phage) integrates into the host genome at a specific attachment site (att) and remains dormant until later point (lysogeny)
How does specialized transduction occur in λ phage?
λ phage integrates into attB sites in E. coli, near gal and bio genes. Abnormal excision can mistakenly include one of these genes in the phage genome.
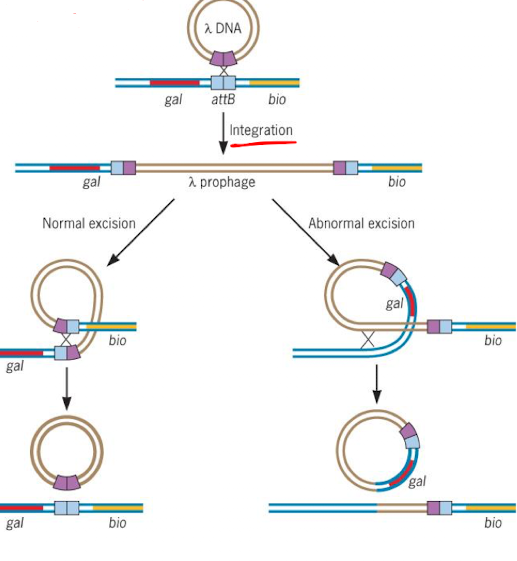
How does the transferred bacterial DNA recombine in specialized transduction?
The phage with bacterial DNA can use homologous recombination to integrate into a new host genome, depending on the number of crossover events.
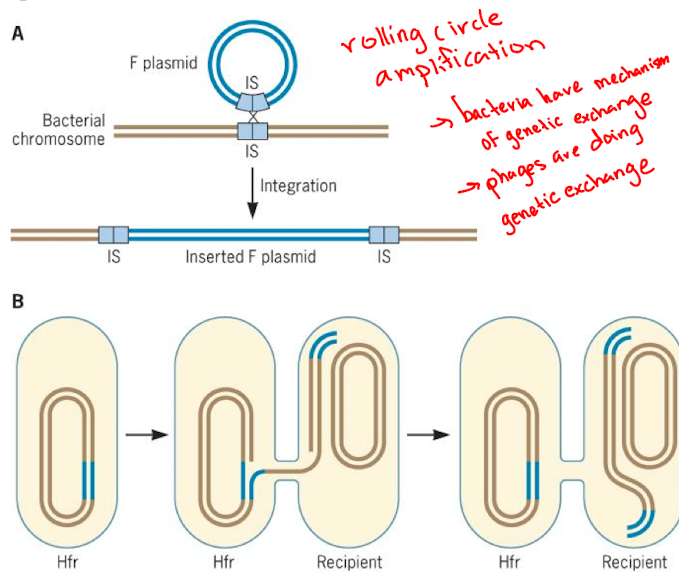
What is bacterial conjugation and what is the key plasmid involved?
Direct transfer of plasmids between bacterial cells; F-plasmid
What structures does F-plasmid encode for conjugation?
Encodes a sex pilus to bring cells together and may encode Type IV secretion system to transfer DNA
How is DNA transferred during conjugation?
Tpe IV recreation system transfers plasmid DNA after it is nicked at the brit and processed by rolling circle replication
What is the F plasmid's role in bacterial conjugation?
Facilitates conjugation and can carry insertion sequences, promoting recombination with the bacterial chromosome to form Hfr strains.
It can transfer integrated plasmids, often bringing host genome segments, and the transfer duration helps map gene positions
Potential applications for cloning
express a gene for study
express a gene in a different organism for beneficial effect
Get gene into a system for mutagenesis
keep gene in a plasmid for stable replication
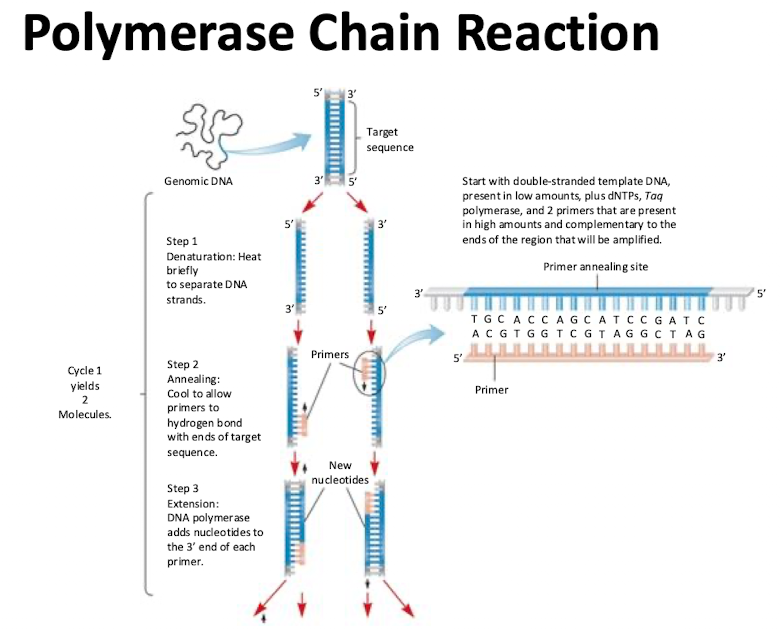
Polymerase Chain Reaction (PCR)
Generates copies of specific DNA sequences in an exponential manner. Each added cycle double the number of copies of the fragment
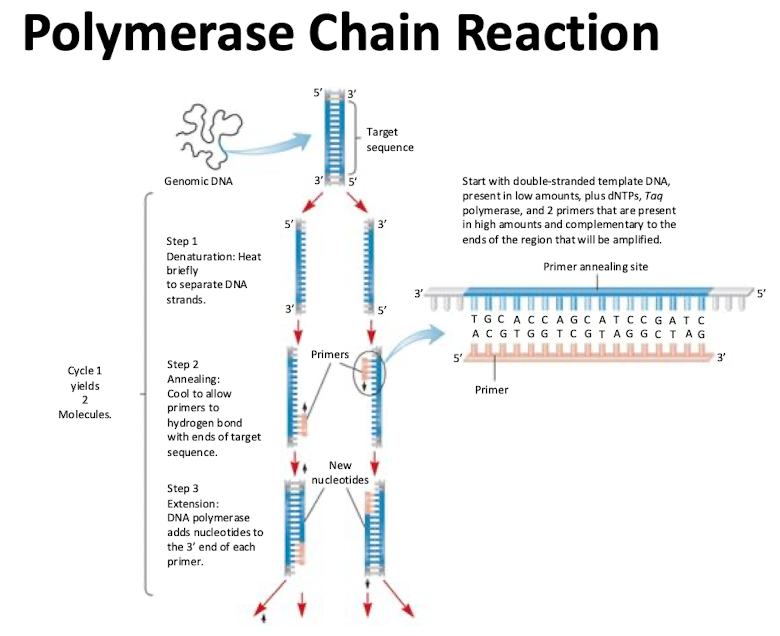
3 Steps to PCR
Denaturing: heat briefly to separate DNA strands
Annealing: cool to allow primers to hydrogen bond with ends of target sequence
Extension: DNA polymerase adds nucleotides to 3’ end of each primer
Restriction Enzymes
protect bacterial DNA by cutting unmethylated foreign DNA
Cut DNA exposes ends, targeting them for degradation
Used for DNA cloning
What is CRISPR-Cas, and how does it function?
bacteria store viral DNA in CRISPR arrays as memory of infections
Cas proteins use stored sequences to recognize and cut invading DNA
Can be used for genome editing
Genome
Organism’s complete collection of heritable information stored in DNA (all of an organism’s DNA)
Genomics
studying the entire genome of an organism
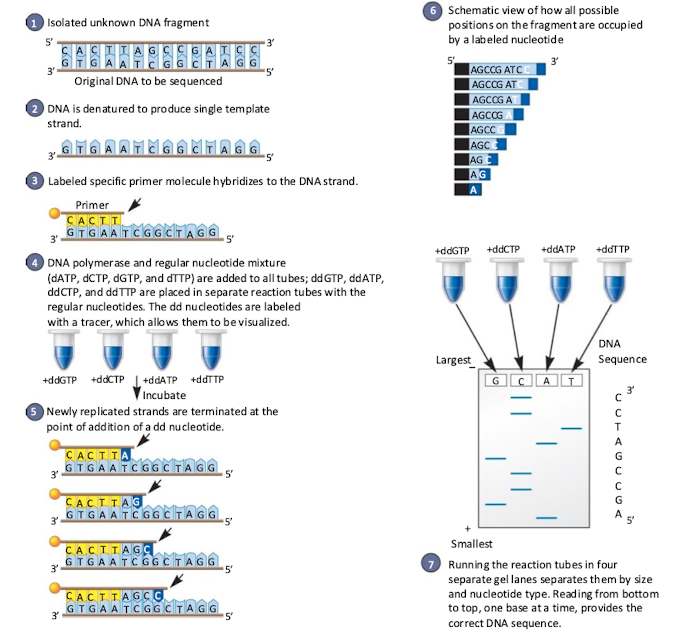
DNA sequencing
Take fragment you want to sequence, add in DNA polymerase and 4 dNTPs. Add radioactive ddNTP in 1 of 4 reactions. ddNTP will stop reaction. (Sanger method)
Good for short sequences but has a variety of problems
Automated Capillary Sequencing
If the labels are fluorescent instead of radioactive, you can run all reactions in one lane.
Downsides to Sanger-style sequencing
long reads but slow and not always accurate
Requires you to fragment genomic DNA and clone into plasmids or bacterial artificial chromosomes (BACs) if you want to sequence a whole genome
Each plasmid has to be sequenced separately
Sequencing by synthesis
uses polymerases and enzymes to create copies of the DNA
Nanopore sequencing
DNA goes through machine reading the electrical current from H bond donors and acceptors
What is genome annotation?
Process of predicting features in a DNA sequence, such as genes, operons, promoter sequences
How is a genome annotation performed?
compare predicted gene sequences to databases of known genes
Identify homologs
find matches to known protein domains
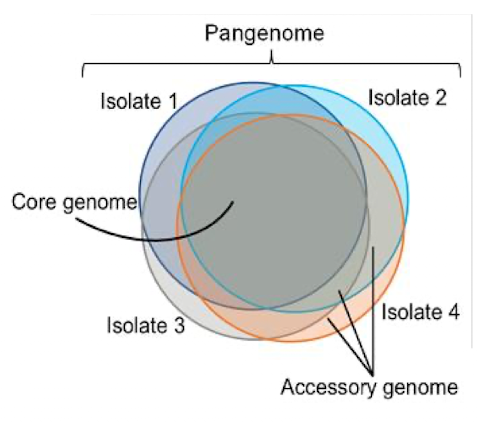
What are the parts to a pangenome?
Core genome: sequences shared between all individuals of the species
Accessory or dispensable genome: partially shared and strain-specific genes (sometimes in plasmids)
Functional Genomics
Diagram of all the cell’s genes products to determine what it can and can’t do
What is metagenomics?
The study of many entire genomes in total
genomics and metagenomics good to study bacteria that can’t be grown in a lab or mixed samples from any environmental source
List 3 applications of genome sequencing
agriculture, energy, disease
Transcriptome
Organism’s complete collection of RNA transcripts
Proteome
organism’s complete collection of proteins. Proteins change according to time, cell type, and cell state
Note: if transcriptomes are changing so are proteomes
genome → transcriptomes → proteome

Metabolome
Organism’s complete collection og metabolites
Can’t get all metabolites with a single method; metabolite is usually defined as any molecule less than 1kDa in size
Order of data to find biological phenotype
Genomics (DNA) → Transcriptomics (RNA) → Proteomics (Proteins) → Metabolomics (biochemical) → Biological Phenotype
Why is morphology not always enough to categorize prokaryotes?
uncultivable organisms
not enough visible differences
What characteristics can be used to classify prokaryotes
Physiological, metabolic, morphological
Strain definition and how to distinguish them (3)
Descendants of a single pure culture
Biovars: strains that are different based on biochemical attributes (metabolic)
Morphovars: strains that look different (colony/cell morphology under microscope)
Serovars: strains that have different antigenic properties
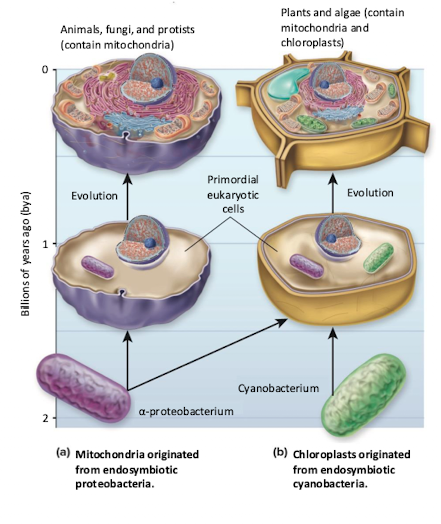
Endosymbiotic Theory
Eukaryotes engulfed bacteria at various points in time and of digesting them, they became stably incorporated into the cell over time
Analysis of genomes’ mitochondria and chloroplast help prove they are descendants of bacterial cells
Thermophilic ancient lineages
look like LUCA
Most deeply-branching lineages
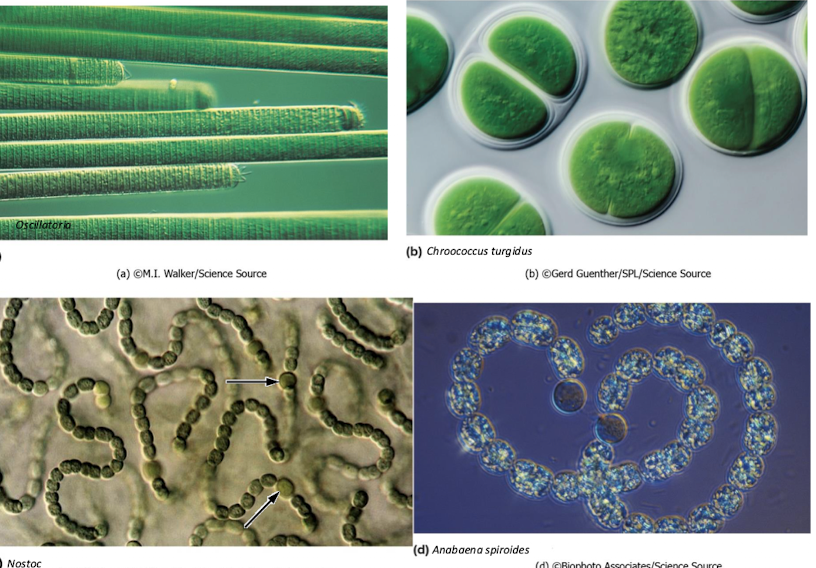
Cyanobacteria
phyletic group
contain only bacterial oxygenic photosynthesizers
can fix nitrogen
some filamentous, some colonial
neat differentiation potential
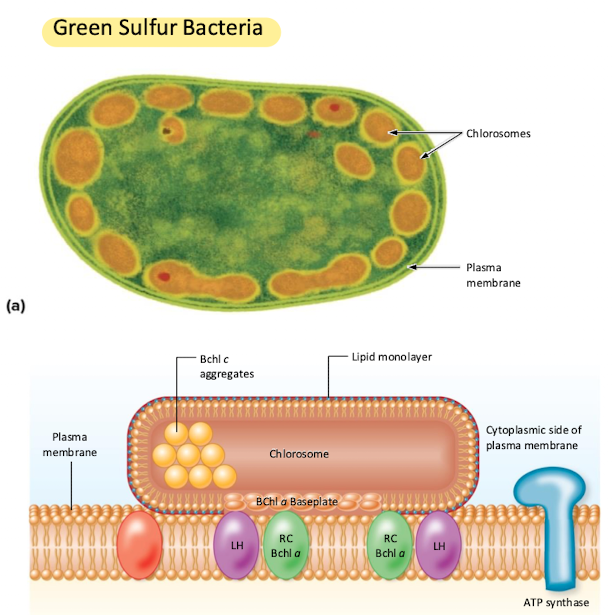
Green Sulfur Bacteria
Chlorobi
Anaerobic photolithoautotrophs (get electrons from inorganic substance using light energy and fix carbon)
Have chlorosomes to contain pigments
Uses reductive TCA to fix CO2
When they oxidize sulfur, they accumulate elemental sulfur on the outside of the cell
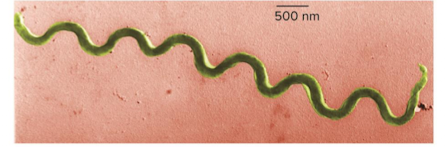
Spirochaetes
Bacterium that “drills” in
Many/most are free-living and anaerobic
Axial filament: an internal flagella that lets the cell move though highly viscous media
hides flagella where it can’t provoke antigenic response