1. Transposition
1/63
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
64 Terms
Transposition is a type of
Recombination
Classic example of transposition in E. coli
integration of λ bacteriophage into the bacterial genome.
attP site
Phage recombination site on the phage genome to facilitate integration
attB site
Bacterial recombination site in E. coli where integration occurs
Lysogenic cycle
After complete phage integration into bacteria genome, the prophage will be replicated along with the bacterial DNA during cell division.
Site specific recombination
is a process by which specific sequences of DNA are recognized and recombined by recombinase enzymes, allowing for the integration or excision of genetic material in a targeted manner.
Recombinase enzymes
recognize certain DNA sequences where recombination occurs, facilitating the integration or excision of genetic material.
Site specific recombination requires ____
short regions of homology for alignment (15-20 bp)
Is site specific recombination only used for insertion?
No, it is used for insertion, excision, and inversion
Site specific recombination- excision
is a process where specific DNA sequences are recognized and removed, allowing for the precise removal of genetic material from a genome.
Site specific recombination- inversion
is a process where a specific DNA segment is flipped orientation within the genome, allowing for the rearrangement of genetic material.
Recombinase enzymatic mechanism
Active site (serine/tyrosine) promotes hydrophilic attack on DNA phosphodiester, then covalent bond conserve energy to use to rejoin DNA strand break
Why does the recombinase enzyme want to conserve that phosphodiester bond break energy in the form of a temporary covalent bond?
Because then no ATP is required
The active site on recombinase enzymes can be which 2 hydrophilic residues?
Serine or tyrosine
Serine recombinase
swap entire double stranded DNA ends through a mechanism involving a transient covalent bond.
Tyrosine recombinase
Performs single stranded breaks and swaps the top strand, then repeats for the bottom strand (so not at the same time)
T/F: Site specific recombination allows exchange between 2 lengths of DNA that are largely unrelated
True, only 15-20 bp of homology are needed
Bonus information: What is the mechanism of diversification for antibody diversification?
Site specific recombination of immunoglobulin genes
Transposable elements
are DNA sequences that can change their position within the genome, often creating or reversing mutations and altering the cell's genome size.
Movement of DNA requires actions of what proteins:
Proteins that bind to recognition sequences on mobile
Enzyme that catalyzes actual movement
Do transposable elements require homology between donor and insertion
No, they can move without requiring homology.
Class I transposons
Retrotransposons
Retrotransposons
Transcribes itself from the original sequence into an RNA copy to move it and reverse transcribe it into other places in the genome.
Class II
DNA transposons
DNA transposons
move by cutting themselves out of the original and inserting into a new location
DNA transposons encode their own
transposase enzyme, allowing them to move within the genome.
Which transposon moves by a copy/paste mechanism
Retrotransposons because they transcribe into an RNA copy to move
Which transposon moves by a cut/paste mechanism
DNA transposons, which excise themselves from one location and insert into another.
2 types of retrotransposons (class I)
LTR
Non LTR
LTR (virus-like) retrotransposon
Has long terminal repeats and encodes integrase and reverse transcriptase
Non LTR retrotransposon
Has a poly A tail with multiple ORFs for RT, endonuclease, and integrase
Conservative transposition
A mechanism of transposition where the transposon is excised from one location and inserted into another, leaving no copy behind. (Class II DNA transpositions)
Replicative transposition
the transposon is replicated and inserted into a new location, resulting in two copies of the transposon in the genome. (Class I retrotranspositions)
Which class of transposon is found in human DNA
mostly Retro transpositions (class I)
Which types of class II retrotransposons are found in humans
LINE and SINE
LINE
Long Interspersed Nuclear Elements (non LTR)
SINE
Short Interspersed Nuclear Elements (non LTR)
Step 1/5 of class II (cut/paste) transposition
First transposase forms a multimer at the inverted repeat region
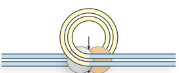
Step 2/5 of class II (cut/paste) transposition
Transposase cleaves 1 strand on each side to expose the 3’ OH ends
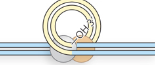
Step 3/5 of class II (cut/paste) transposition
The free 3’OH groups attack the other strand to make the hairpin structure and excise the segment from the donor
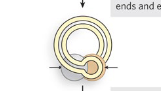
Step 4/5 of class II (cut/paste) transposition
Transposase cleave the hairpin to reexpose 3’ OH ends, which then attack target DNA
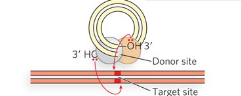
Step 5/5 of class II (cut/paste) transposition
The transposon is integrated into the target DNA, resulting in a double-strand break that is repaired by DNA Pol and ligase
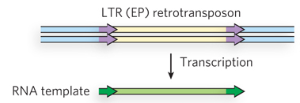
Step 1/5 of class II (copy/paste) LTR retrotransposon
Transcription of retrotransposon by host machinery (encodes RT and integrase) to produce RNA template
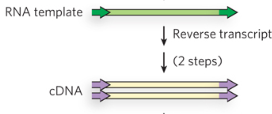
Step 2/5 of class II (copy/paste) LTR retrotransposon
Reverse transcription of the RNA template into DNA, producing a cDNA of retrotransposon.
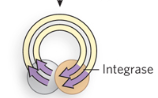
Step 3/5 of class II (copy/paste) LTR retrotransposon
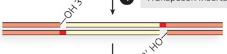
Integrase binds to LTR on both ends of the retrotransposon to circularize cDNA
Step 4/5 of class II (copy/paste) LTR retrotransposon
Free hydroxyl ends on circular cDNA attack target DNA
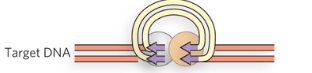
Step 5/5 of class II (copy/paste) LTR retrotransposon
Integration by recombination
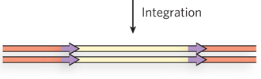
Step 1/6 of class II (copy/paste) NON LTR retrotransposon
transcription of the full-length retrotransposon by
host transcription machinery; ORF 1 and 2 encode RT,
endonuclease, and integrase
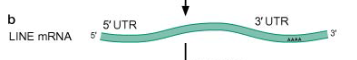
Step 2/6 of class II (copy/paste) NON LTR retrotransposon
ORF1 and ORF2 proteins (RT, endonuclease, and integrase) will bind to transposon RNA
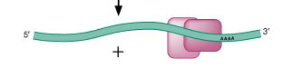
Step 3/6 of class II (copy/paste) NON LTR retrotransposon
Poly A tail guides transposon to target DNA and anneals to form DNA/RNA hybrid
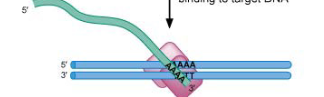
Step 4/6 of class II (copy/paste) NON LTR retrotransposon
Endonuclease will cleave target DNA to produce free 3’ OH
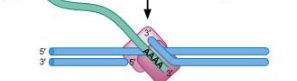
Step 5/6 of class II (copy/paste) NON LTR retrotransposon
poly-t sequence of target DNA serves as template for RT activity to generate cDNA of retrotransposon
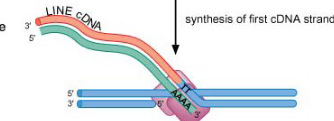
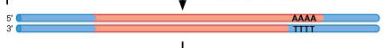
Step 6/6 of class II (copy/paste) NON LTR retrotransposon
cDNA is integrated by integrase and repaired
Autonomous transposons
Encode all necessary parts for transposition like transposase and integrase, so they move independently
nonautonomous transposons
Require assistance from autonomous transposons for transposition, as they lack the genes needed for movement.
Do transposons contribute to evolution
Yes, they can create genetic diversity by facilitating gene rearrangements, recombination’s, and mutations.
Exon shuffling
is a process where exons from different genes are mixed and matched during recombination, leading to new gene combinations and potentially new functions.
“unclean transposition” results in what phenomenon
Exon shuffling
Endogenous retrovirus (ERV)
endogenous viral elements in the genome that
closely resemble and can be derived from
retroviruses
ERVs are crucial for what specific process in mammal reproduction
Placenta development and birth timing because it needs the retrovirus envelope protein syncytin-1 AND CRH hormone
Syncytin-1
a protein derived from an endogenous retrovirus that plays a key role in the formation of the placenta by facilitating the fusion of trophoblast cells.
CRH hormone
corticotropin-releasing hormone, linked to the control of birth timing in humans
ERV role in digestion
ERV activity duplicates amylase gene, which efficiently digests starch
How would exogenous retroviruses have evolved from LTR transposons
Gain of envelope, allowing them to survive and replicate outside the host cell.