ch. 13 the molecular basis of inheritance
1/47
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
48 Terms
Rosalind Franklin (1952)
scientist who studied the DNA molecule
developed an x-ray image of the DNA Helix
not credited for her work
enabled Watson and Crick to also study DNA molecule
James Watson and Francis Crick (1962)
scientists who were credited with the structure of DNA and researched the pairing of nitrongenous bases (who pairs with who)
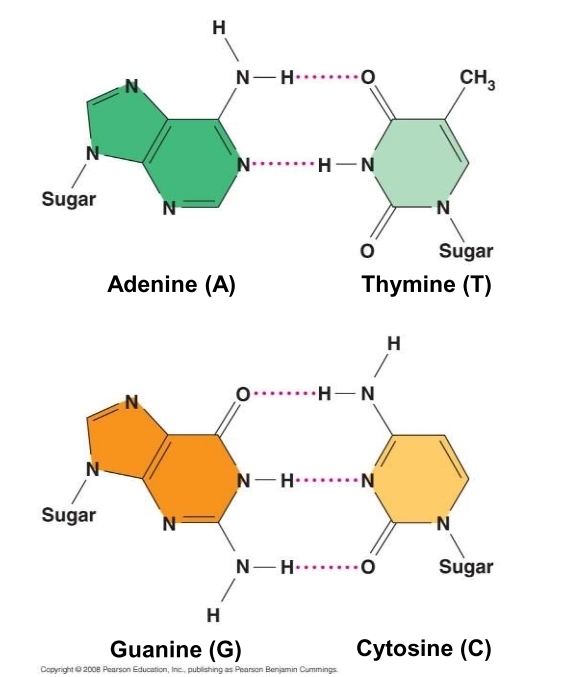
how are nitrogenous bases paired?
purine (A & G) with purine (T & C)
Matthew Meselson and Franklin Stahl
scientists that studied DNA replication and that the two new strands are half old and half new
semiconservative
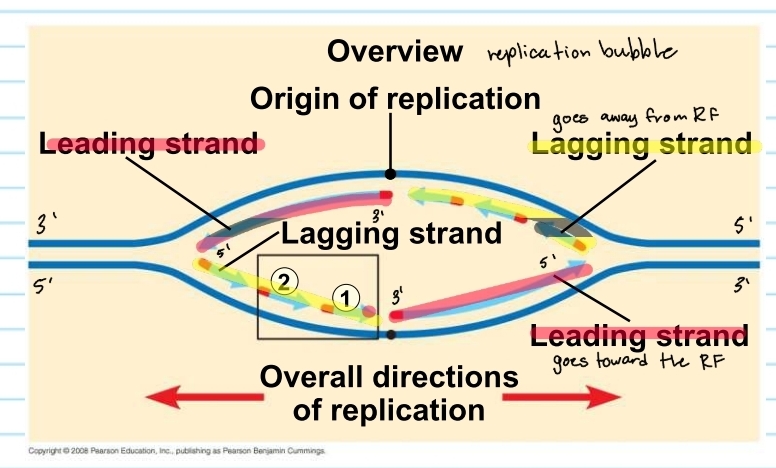
origins of replication
where the two strands are separated
opens up a replication bubbles
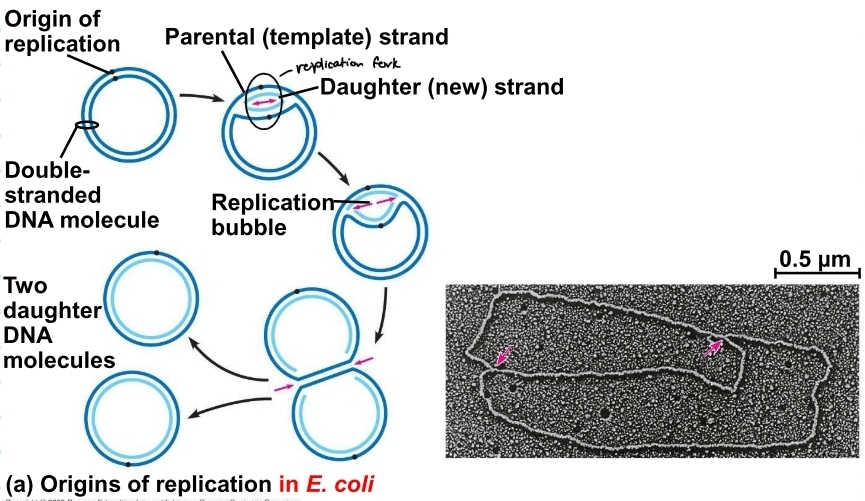
replication bubble in prokaryotes
based off of ecoli
only has one replication bubble
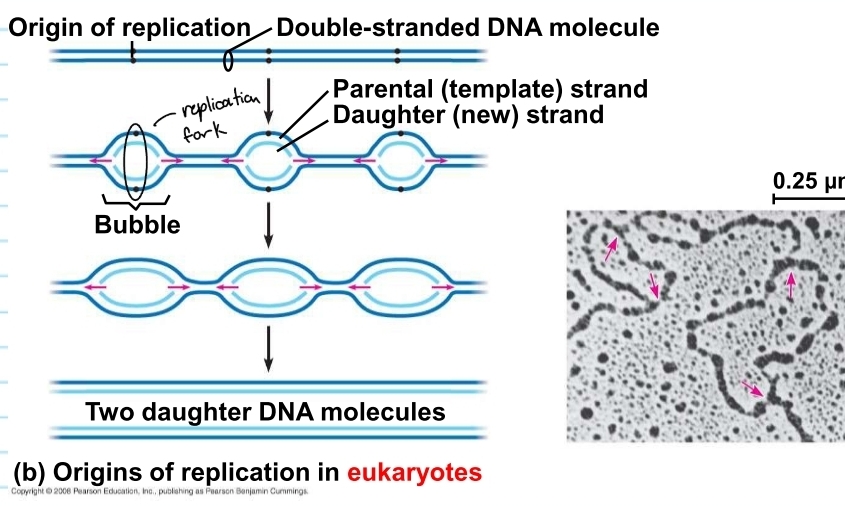
DNA replication in eukaryotes
based on plants and humans
has multiple replication bubbles
these eventually stretch out and combine
DNA polyermase and elongation
enzymes that add nucleotides only to the free 3’ end.
meaning the new DNA strand can only elongate in the 5’-3’ direction
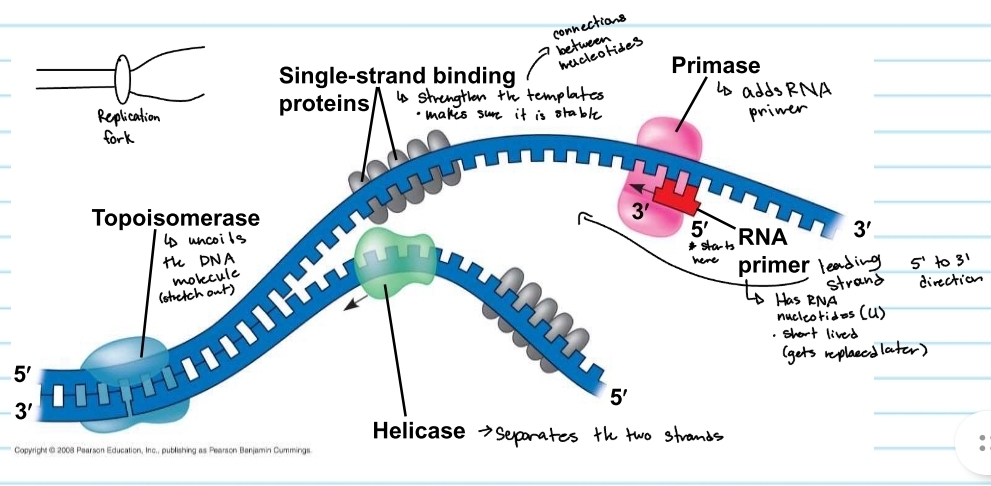
Helicase
separayes the two strands
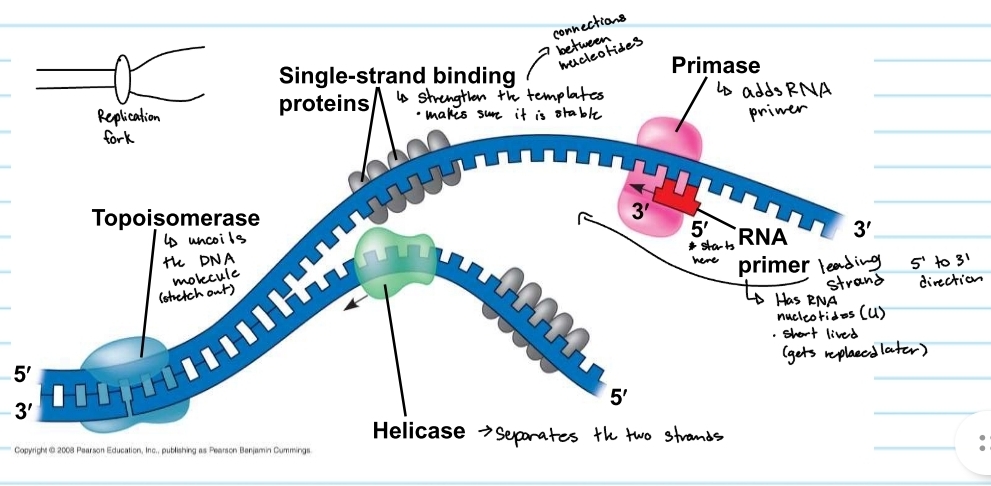
topoisomerase
uncoild the DNA molecule (stretch out)
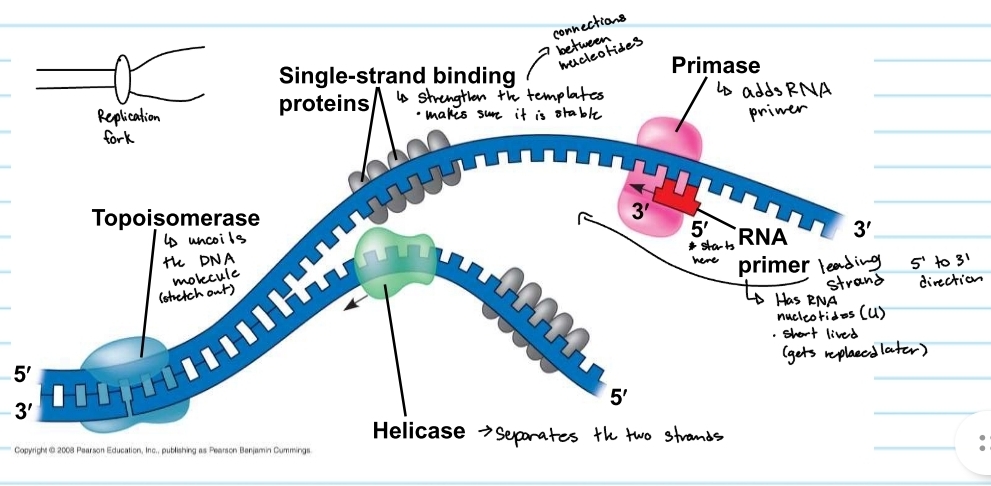
single-strand binding proteins
strengthens the templates
keeps them stable
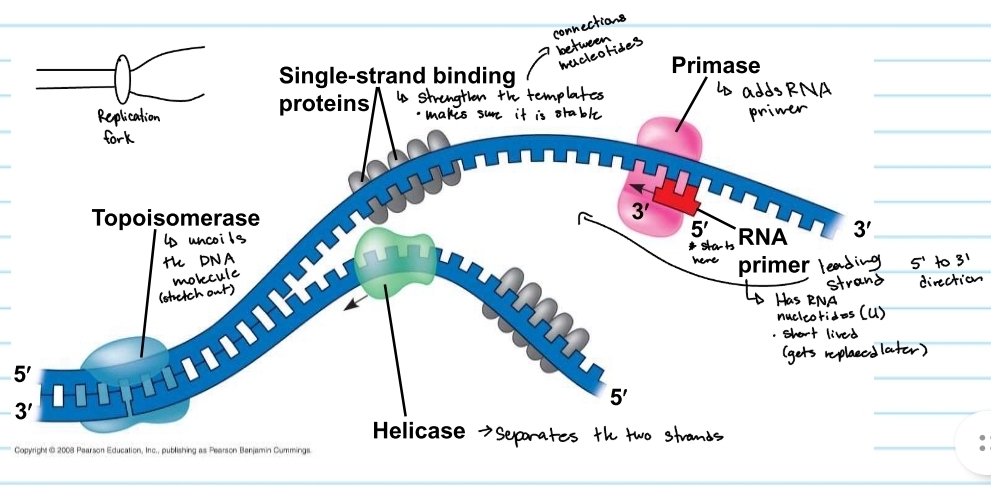
primase
adds the RNA primer
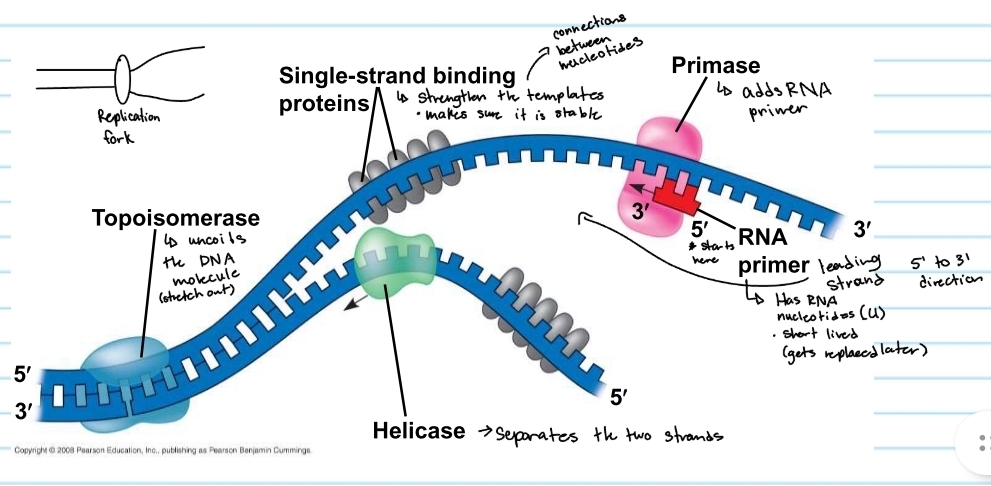
RNA primer
shows where DNA polymerase will start
made of RNA nucleotides (no thymine)
gets replaced later (short lived)
DNA Polymerase 3 and 1
polymerase 3
the enzyme that adds nucleotides and synthesizes the new strand
polymerase 1
enzyme that replaces RNA primers with DNA nucleotides
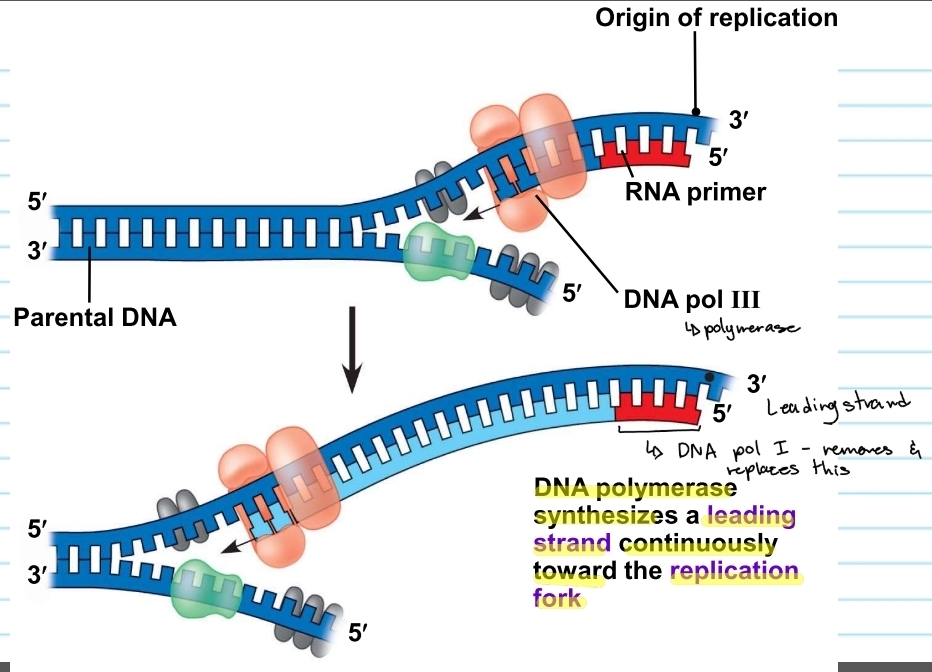
leading strand
the template that is 3’ to 5’
DNA polymerase will synthesize CONTINUOUSLY replication fork
5’ to 3’
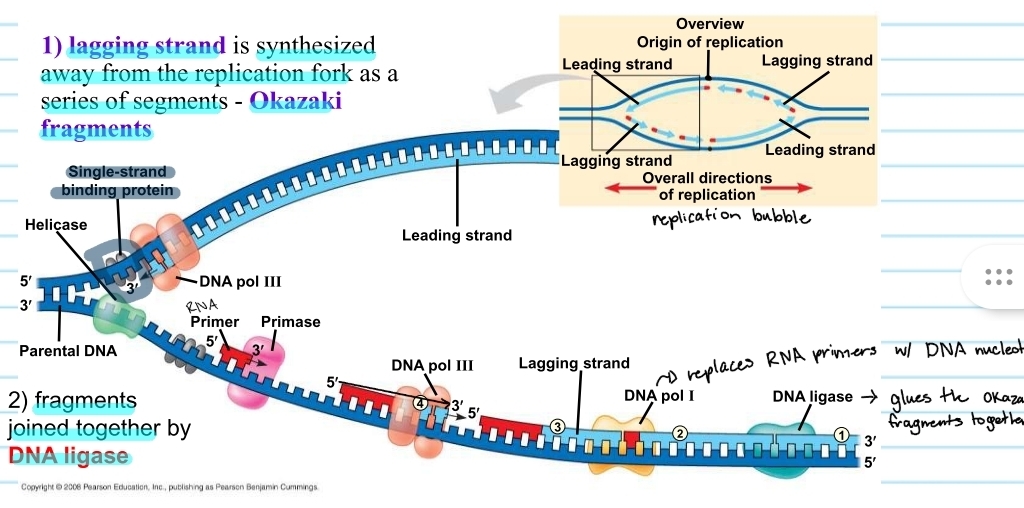
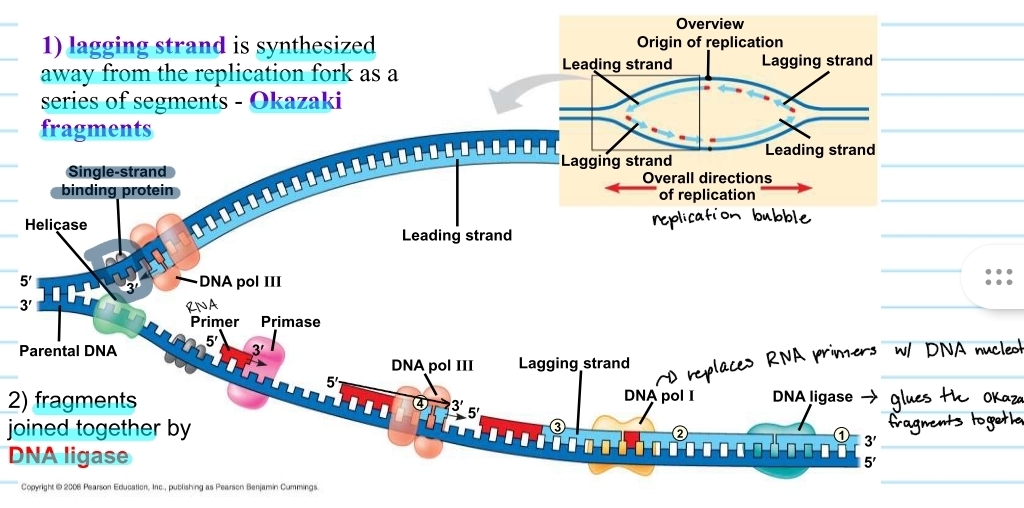
lagging strand
the template that is 5’ to 3’
DNA polymerase must follow a 5’ to 3’ direction, so the new strand for this template is synthesized in chunks away from the replication fork
chunks are known as okazaki fragments
RNA primers must be placed each time
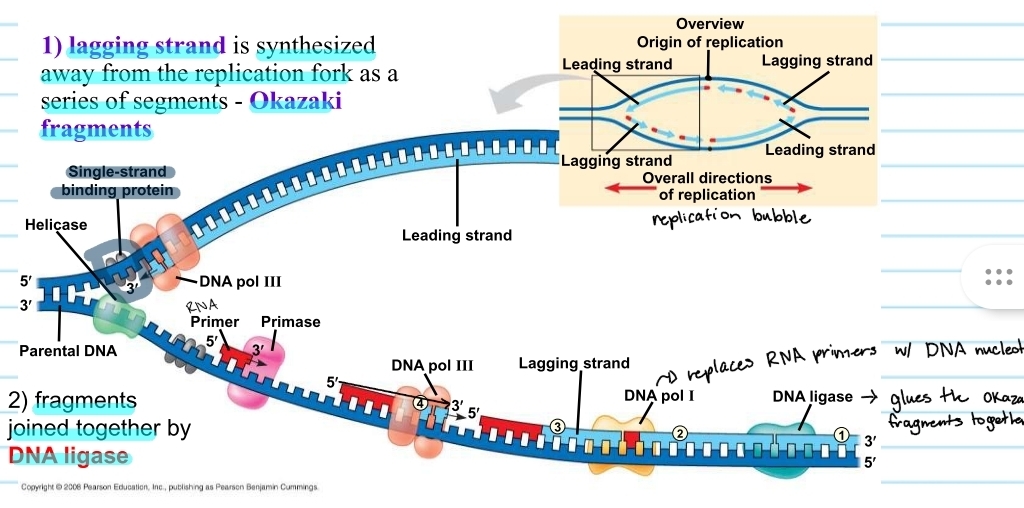
DNA ligase
the enzyme that glues the okazaki fragments together
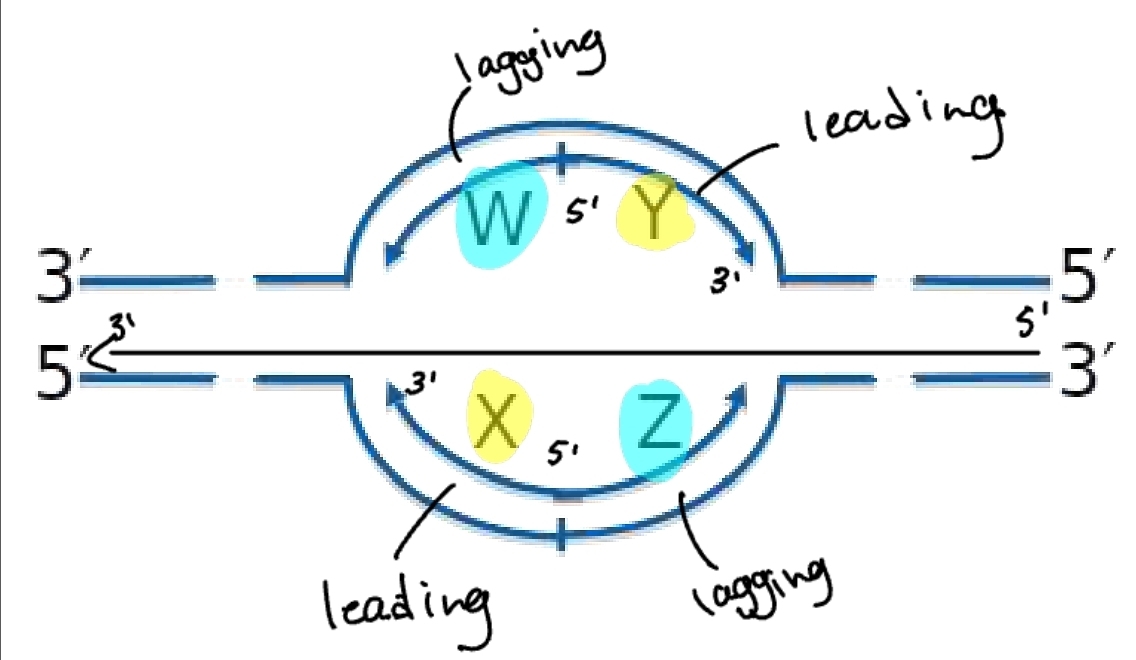
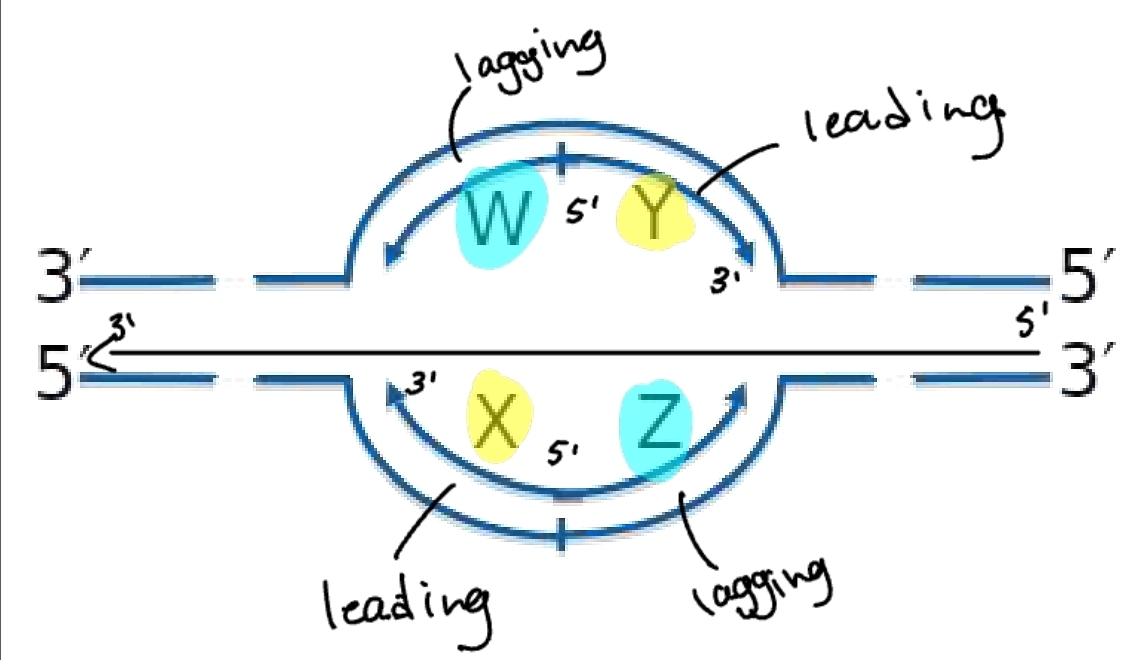
replication bubble - lagging and leading strand
from the origin of replication the lagging strand and leading strand alternate
this is because it depends which template it is on and which direction it is going
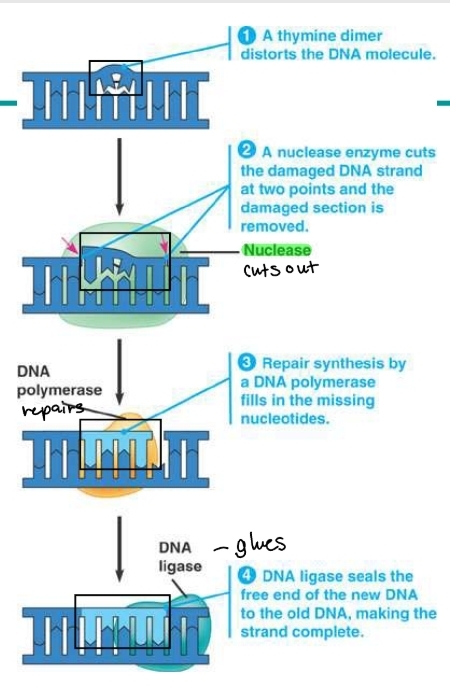
which enzyme proofreads and repairs DNA?
DNA polymerase
the enzyme corrects eerors in base pairing
e.g. a thymine dimer (connection between nucleotides in the wrong way
polymerase checks for the error
a nuclease enzyme then cuts out the damaged DNA section
DNA polymerase repairs the cut out section with the missing nucleotides
DNA ligase will then glue the new section with the nucleotides around it
xeroderma pigmentosum (XP)
a disorder caused by an inherited defect in the nucleotide excision repair
disorder occurs when damage caused by ultraviolet light that does not get corrected
individuals with XP are hypersensitive to sunlight
children who have XP can develop skin cancer (melanoma or basal carcenoga) by age 10 (moonlight kids)
mostly affects the eyes and areas of skin exposed to the sun
follow autosomal pattern of inheritance and effects about 1 in 1 million people in the U.S., but can be much higher in certain parts of the world
what does a chromosome consist of?
a DNA molecule that is packed together with proteins
bacterial chromosome
a double-stranded, circular DNA molecule associated with a small amount of protein
in bacterium, the DNA is super coiled and found in a region called the nucleoid
eukaryotic chromosomes
have linear DNA molecule associated with a large amount of protein
these proteins are called histones
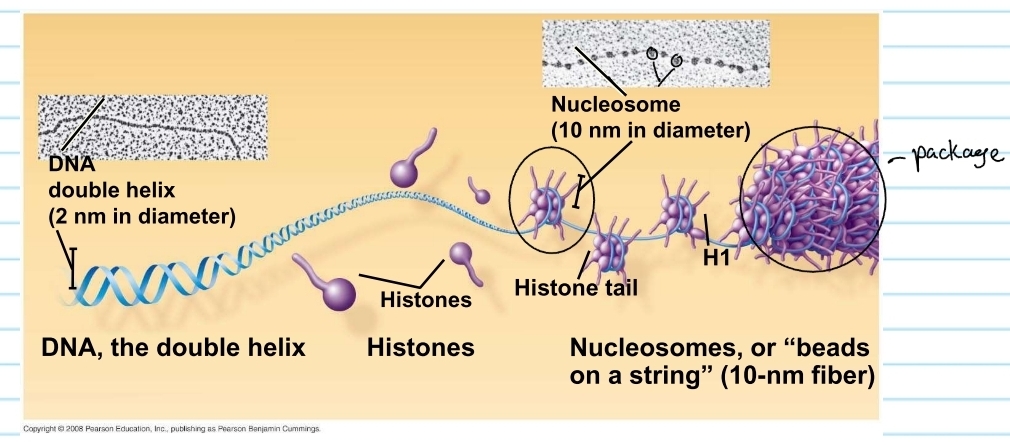
histones
proteins that are responsible for the first level of DNA packing in chromatin
it super coils the DNA
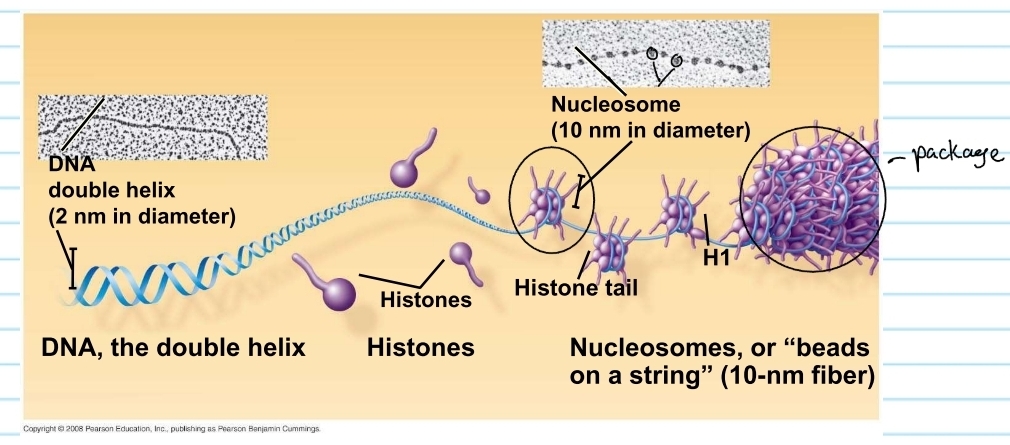
nucleosome
consists of DNA wound twice around a protein core of 8 histones
these pack together into a chromosome (chromatids)
Frederick Griffith (1928)
scientist researched two types of Streptococcus pneumoniae (bacterium)
Pathogenic (S - smooth)
Harmless (R - rough)
concluded the Transformation factor
pathogenic (S cells)
have capsules that protect them from the animals’ immune system
surrounded by protective coating known as smooth cells
causes the virus
harmless (R cells)
bacteria that do not have capsules
no protective coating
does not cause the virus because the immune system will kill it
harmless and known as ROUGH cells
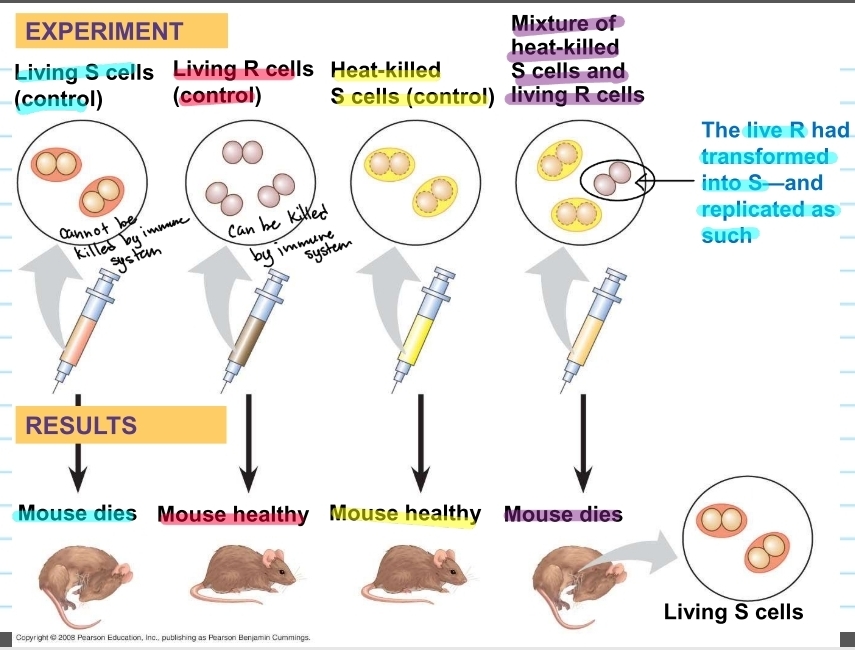
Griffith's experiment
gave mice different bacteria cells
mouse given S cells (control) → died
mouse given R cells (control) → lived (healthy)
mouse given heat-killed (killed by high temps; cannot work) S cells (control) → lived (healthy)
mouse given a mixture of heat killed S cells and living R cells → died (found living S cells in the mouse)
concluded R bacteria, that normally be killed, transformed into S bacteria via an unknown, heritable substance (factor)
transformation
change in genotype and phenotype due to assimilation of foreign DNA by a cell
S genotype got assimilated into R genotype (tranformed)
later concluded that DNA was the heritable factor that would cause tranformation
what does this show about viral DNA
additional evidence that DNA was the genetic material came from studies of virus that infect other bacteria
such as bacteriophages (or phages)
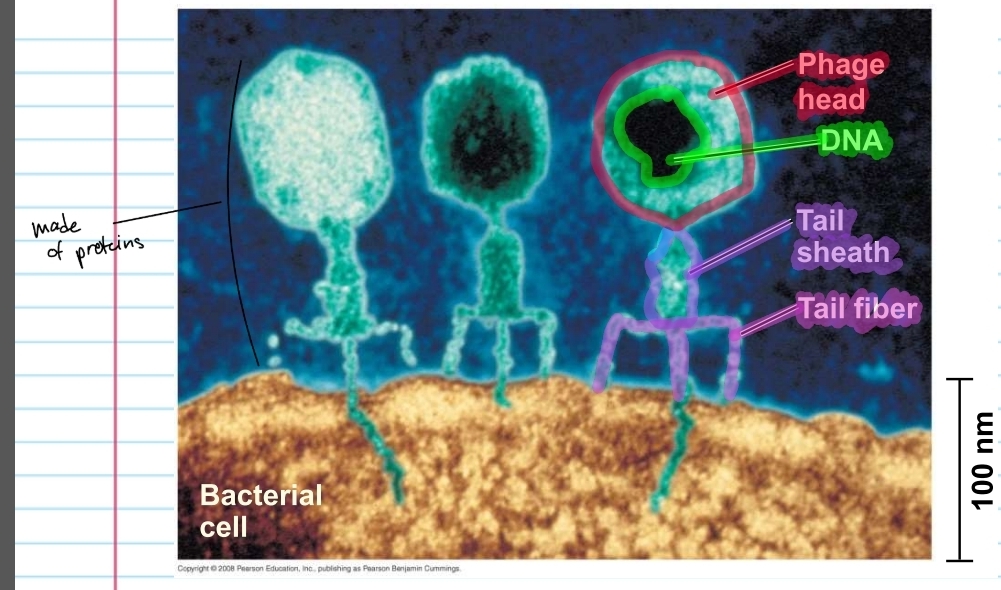
phage
consists of a phage head with DNA inside, a tail sheath and tail fiber
everything except the DNA is made of proteins
how do phages reproduce?
a phage will attach to a host cell's plasma membrane (with the fibers) and inject the DNA, leaving the protein behind
the DNA will enter the host cell, while the original DNA gets destroyed
the enzymes and nucleotides in the host cell begin to replicate the phage DNA
other enzymes and ribosomes will manufacture phage proteins then assemble to form phages
a phage enzyme will then digest the bacterial cell wall and cause the cell to rupture (lysis)
this lets the formed phages out, can be up to 200 phages, that will go to infect other cells
Alfred hershey and Martha Chase (1952)
researched T2 phages and whether or the protein carries the genetic info or the DNA does
done with an E. coli host + T2 phages
concluded that DNA is the genetic material that is responsible for hereditary
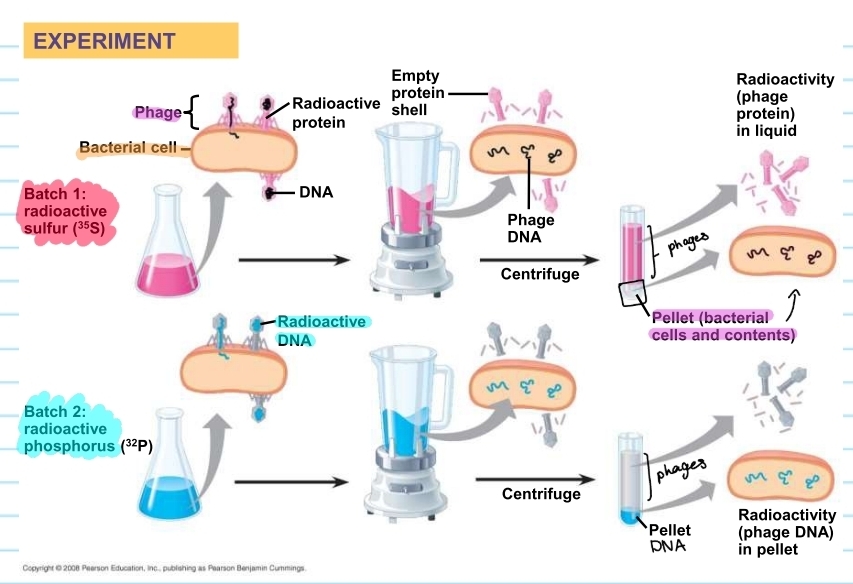
phage experiment process
phages were laced with two different radioactive solutions
one will be on the protein (pink)
one will be on the DNA (blue)
this is to check which provides the genetic info
the pink one showed that the proteins stayed out of the host cell
the sample from the host cell showed no pink pigment
the blue one showed that the DNA went into the cell
the sample from the host cell showed blue pigment
what did the experiment conclude?
DNA is the genetic material that causes hereditary
injected DNA provides genetic info that makes the cell produce new T2 DNA and proteins
Erwin Chargaff (1950)
scientist that studied the DNA composition
varies from one species to the next
developed Chargaff's rule
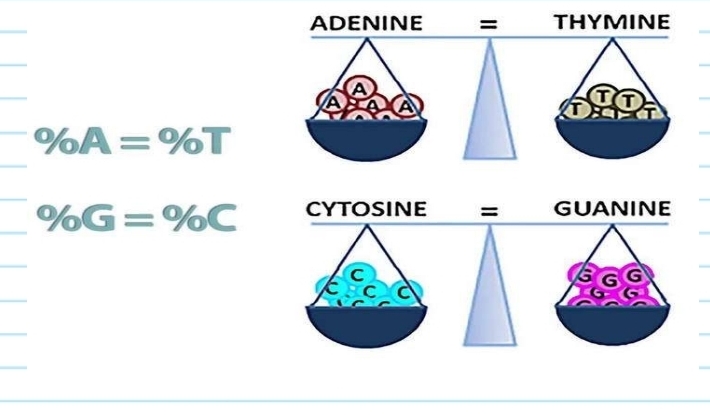
Chargaff's rule
in any species there is an equal number of A and T bases, and an equal number of G and C bases
Maurice Wilkins
scientist who worked w/ and took the DNA picture
received nobel prize with Watson and Crick
Linus Pauling
proposed a three-stranded model of DNA
this was an incorrect model of DNA
DNA cloning
yields multiple copies of a DNA segment
uses bacteria and their plasmids
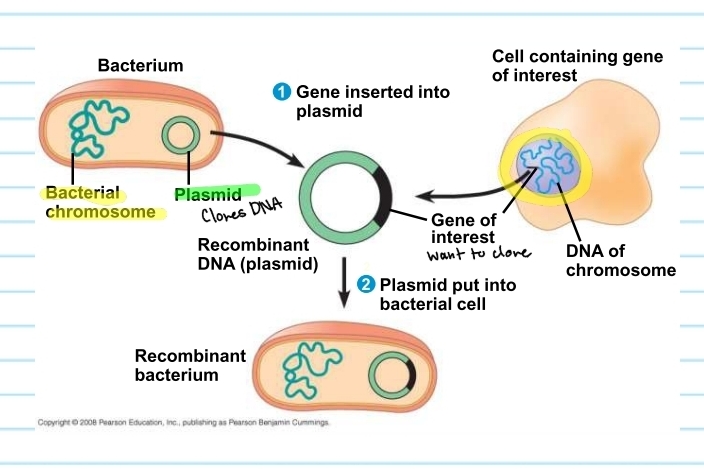
plasmids
small circular DNA molecules that replicate separately from the bacterial chromosome
found in bacterial cells
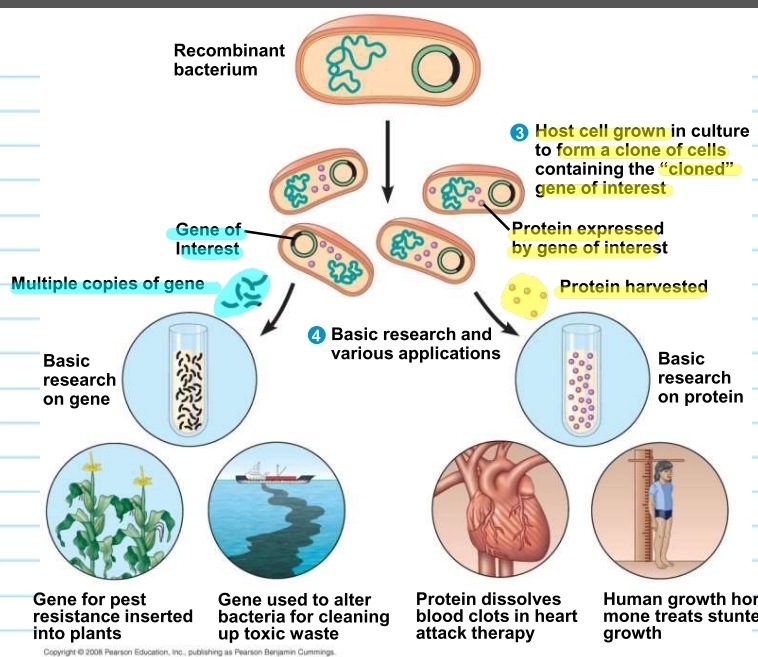
what are plasmids used for?
to create more of the same gene or more of the protein produced by the gene
how do plasmids clone a gene of interest?
plasmid from a bacterium has a gene of interest inserted into it
creates a recombinant plasmid (DNA)
plasmid will then be placed back into the bacterium
this will allow the gene of interest to be cloned
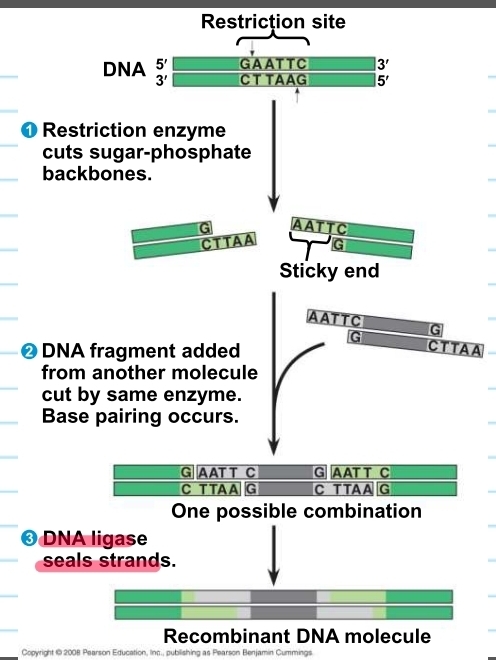
restriction enzymes
used to make recombinant DNA
they cut DNA molecules at a specific DNA sequence called restriction sites
cuts the DNA that will have “sticky ends” that will bond with complementary “sticky ends" of other fragments (glue w/ glue)
DNA ligase seals the bonds betwen restriction fragments (super glue)
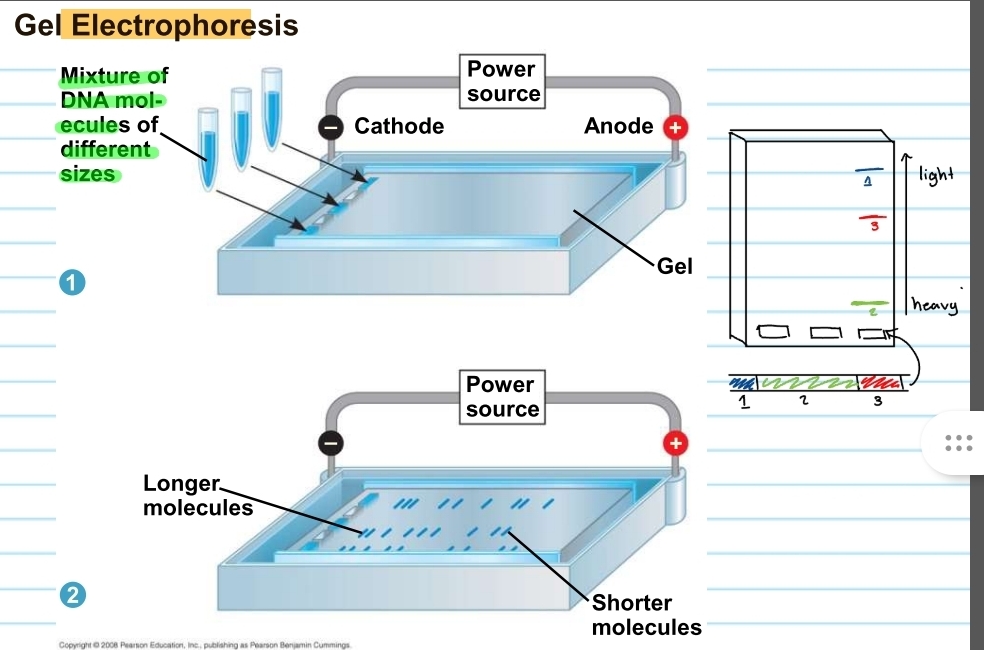
gel electrophoresis
used to sort DNA molecules/fragments that were produced by restriction enzymes
restriction fragment analysis
can also be used to get for diseases such as sickle cell
also used for comparing forensic evidence
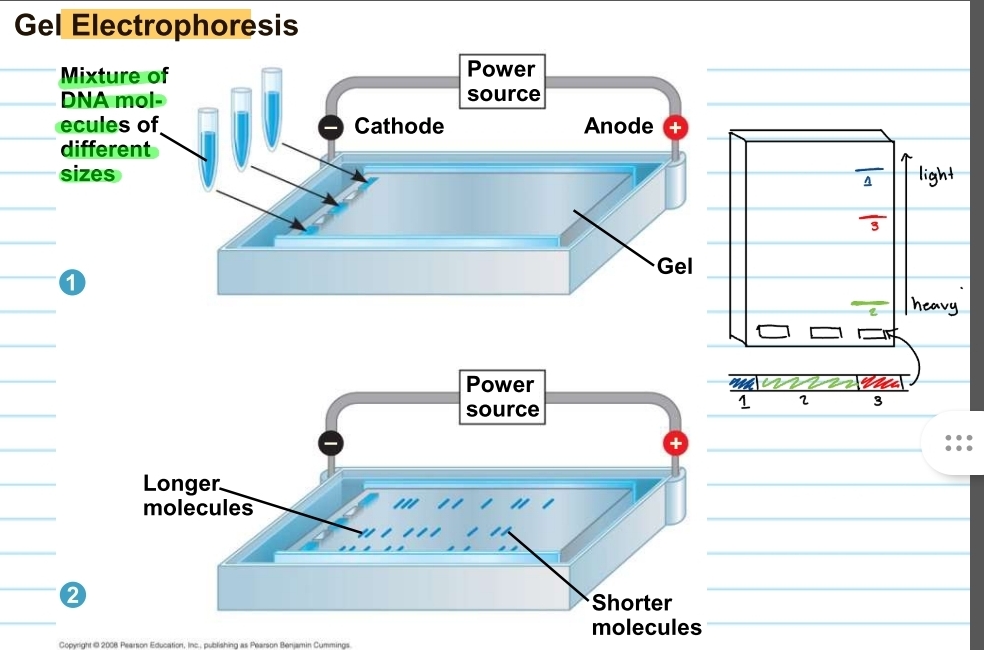
how does the gel electrophoresis work?
consists of a gel and a power source (a cathode end and an anode end)
DNA fragments will be placed into the wells on the cathode side of the gel
these will run depending on their size
fragments closer to the anode end is very light and fragments closer to the cathode end is heavy
fragments are like unique fingerprints
to observe the fragments the gel must be put in UV light
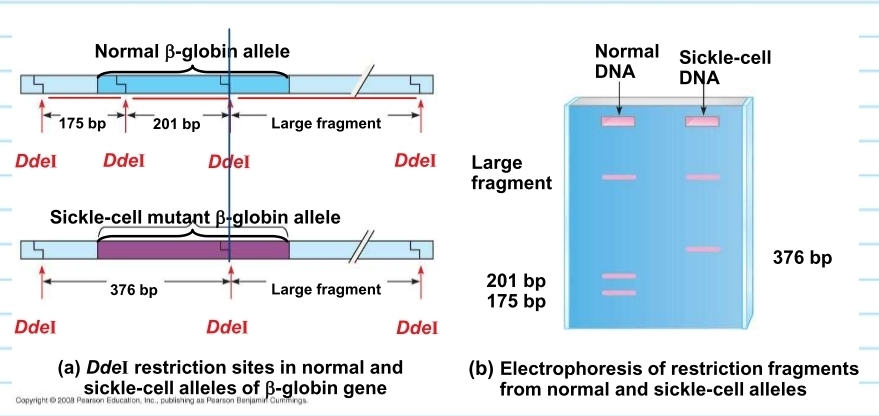
sickle cell DNA - restriction fragment analysis
when observed in gel electrophoresis:
one of the restriction sites (fragments) is destroyed that comes from the DDeI enzyme
as a result the enzyme will generate different fragments when sickle cell DNA is present (will differ from normal DNA)
the fragment in sickle cell DNA combines 2 of the 3 fragments in normal DNA into one fragment
normal DNA has 3 fragments
sickle cell DNA has 2 fragments
this can be observed in a gel electrophoresis