DNA damage and repair L3
1/33
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
34 Terms
DNA Damage Repair
•If DNA damage can be detected quickly, the cell has a chance to repair it
-checkpoints!
Causes of DNA Damage
- endogenous - spontaneous, natural cause,
- exogenous - exposure to an external physical or chemical agent
endogenous
- Mistakes in replication
•Spontaneous physical & chemical events
•
tautomers
altrnaive forms of the same molecule - can interchange
- H atoms on bases can transiently and spontaneously change position
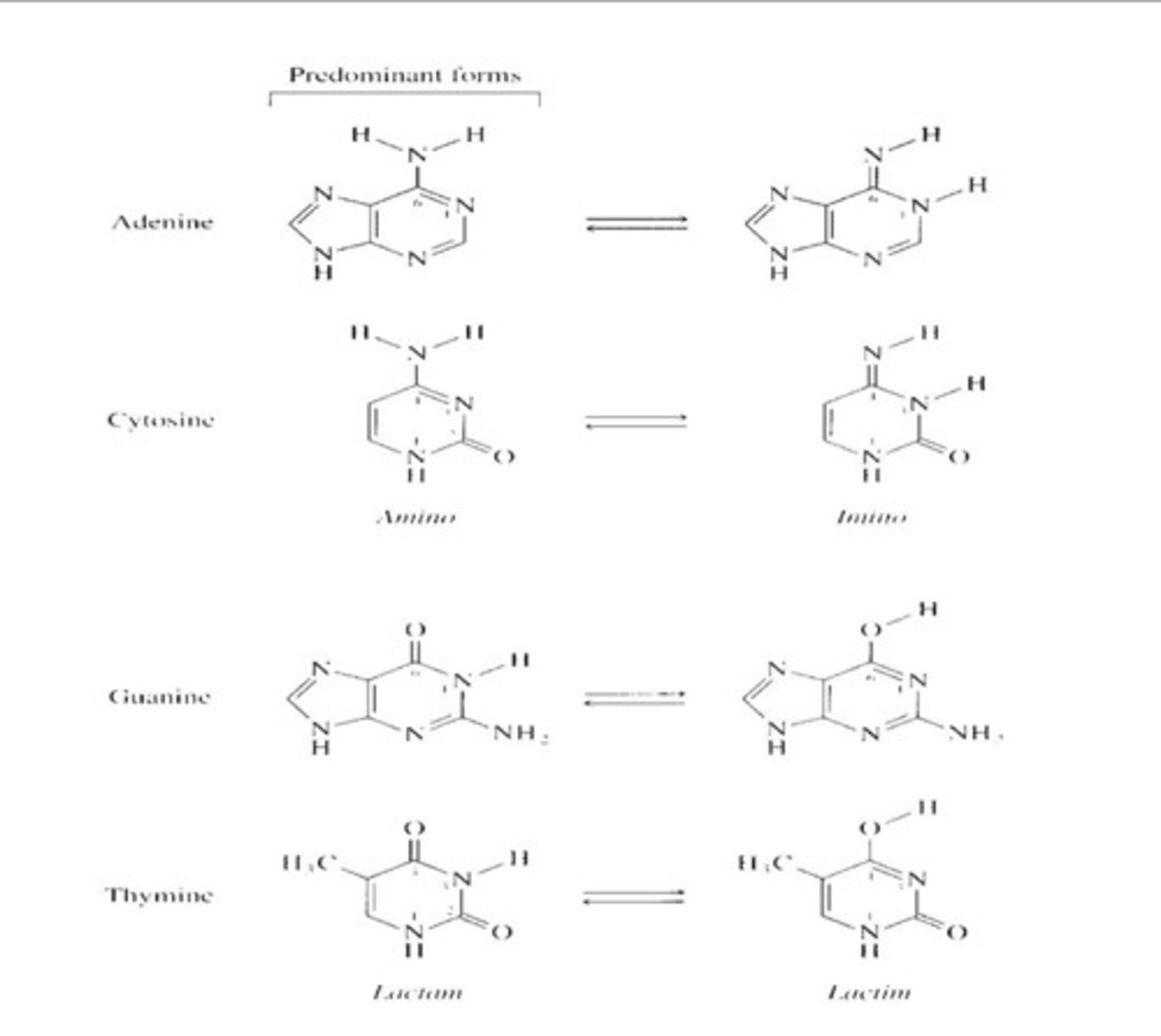
Cant have base pairing to tautomer - 2 H cant bond will bond to will instead bind to a imino form of nucleotide which causes mutations
Tautomers: Base-pairing
Non-Watson Crick Base Pairing:
Non-complementary base pairing
A* = C
G* = T
T* = G
C* = A
molecules within cells are constantly colliding -
brownian motion
Collisions with DNA can remove purine base from nucleotide (hydrolysis of glycosidic bond).
Phosphate-Sugar backbone left intact.
causes deletions by depurination
Apurinic (AP) site
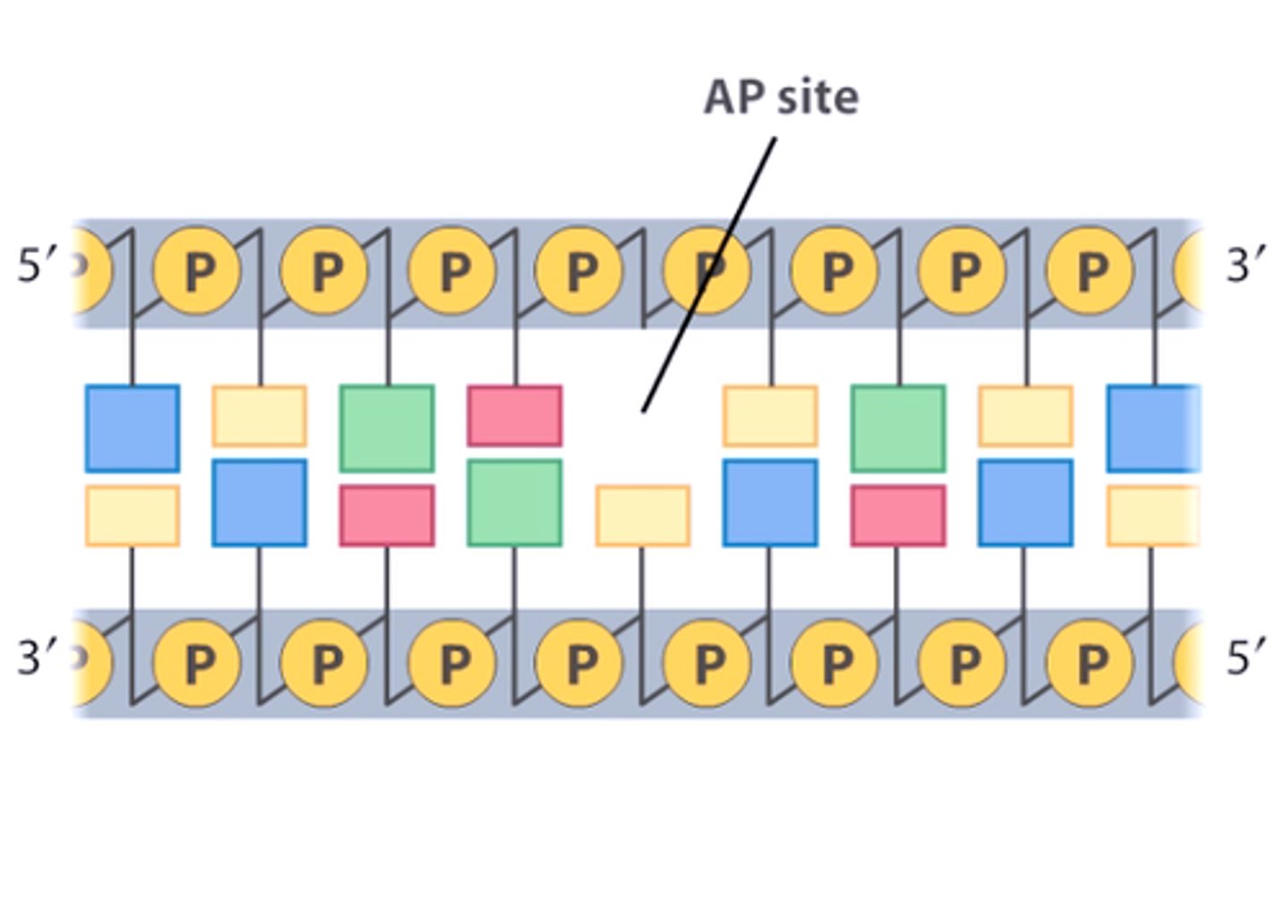
depurination
•AP site causes polymerase to stall during replication.
•Can cause deletion mutation.
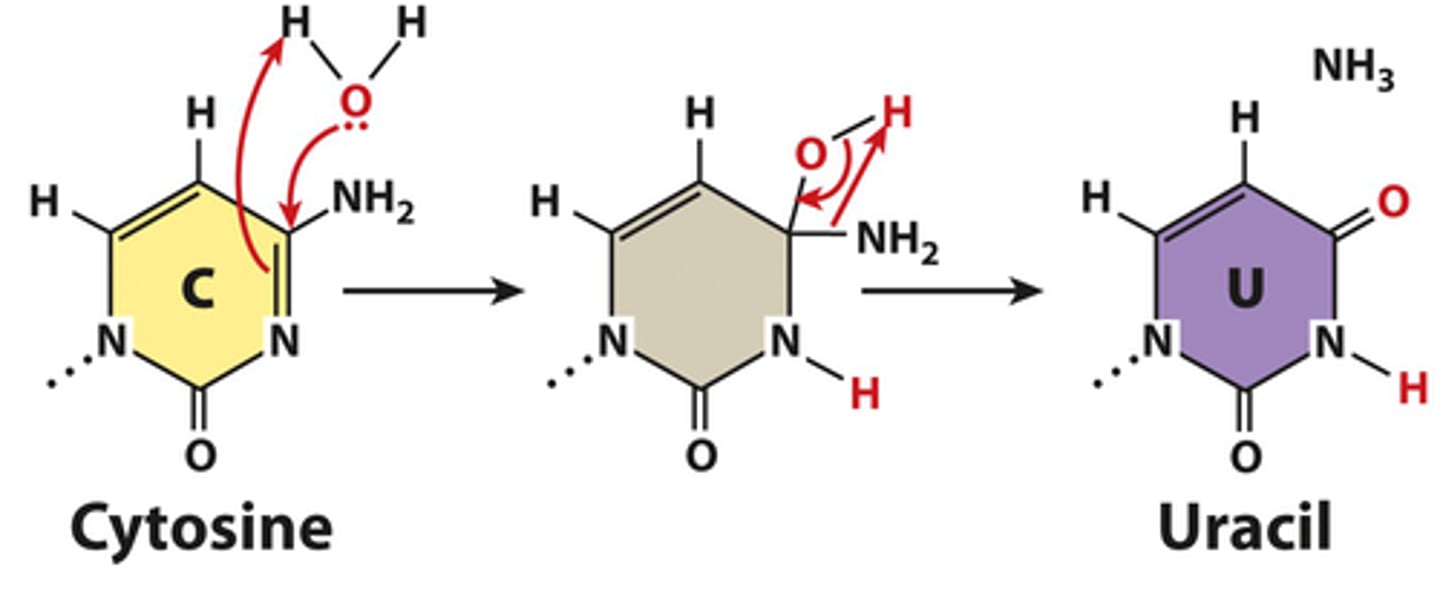
deamination
•Removal of amino group from base.
- causes substitution mutation - cystine to uracil
- C pairs with G
- U pairs with A
MUTATION IS MADE PERMANENT BY REPLICATION
Repair mechanisms MUST act prior to replication being completed
Overall Strategy -
Recognise
Remove
Seal the gap
-Direct Reversal Systems - proofreading
-Excision Repair Systems
-Recombination
-Excision Repair Systems
Performed if damage occurs only on one DNA strand.
The aim is to restore the original sequence using the other strand as a template.
1.Mismatch Repair (MMR)
Watson-Crick base mismatch immediately after DNA synthesis.
All use at least 2 proteins:
•one to detect the mutation
•one to cut out the DNA
•Sometimes a mediator also
Mismatch Repair E Coli example
•MutS recognises
•MutL recruits the endonuclease
•MutH cleaves the phosphodiester backbone
Exonuclease (MutH)
•removes the mismatched segment of DNA
DNA pol III
synthesises a new segment of DNA using the opposite strand as template
•A way to differentiate between old and new strand
•Methylation of DNA occurs over time following synthesis
- •Mismatch repair systems recognise hemimethylated DNA on old strand
-
proteins in the order in which they participate in methyl-directed mismatch repair.
1. MUT S
2. MUT L
3. MUT H
In methyl-directed mismatch repair, the strand that is repaired is
the non-methylated strand. - new strand
1.Base Excision Repair (BER)
Small, non-bulky lesions - specific glycosylases.
- •Remedy for depurination & deamination
1. Glycosylase hydrolyses
1.Glycosidic bond between deoxyribose and the base
2. AP endonuclease creates
a nick in the phosphate-sugar backbone
3. Deoxyribose phosphodiesterase removes
the remaining deoxyribose phosphate unit
4. DNA pol I inserts
a new nucleotide
5. DNA ligase
seals the phosphodiester backbone
chemical mutagens •Grouped based on mechanism of action:
1.Base Analogues - bases similar to normal DNA bases
●
2.Base Modifying Agents - modify chemical structure and properties of bases
●
3.Intercalating Agents - insert between adjacent bases in DNA
1.Base Analogues
1)Analogue - structural derivative of a parent compound that often differs from it by a single element.
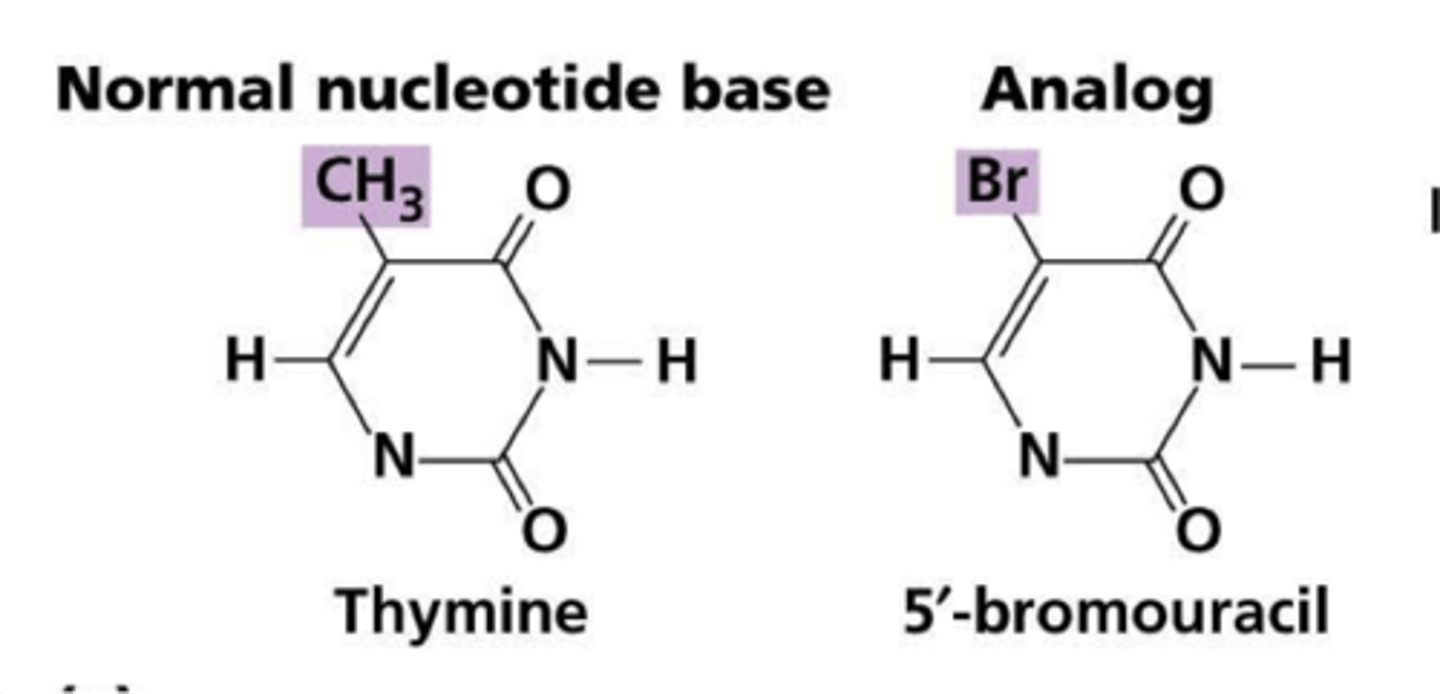
Base-Modifying Agents
Chemicals which modify bases by adding or removing functional groups, changing their base-pairing properties
- e.g.
Hydroxylation
Alkylation
Deamination
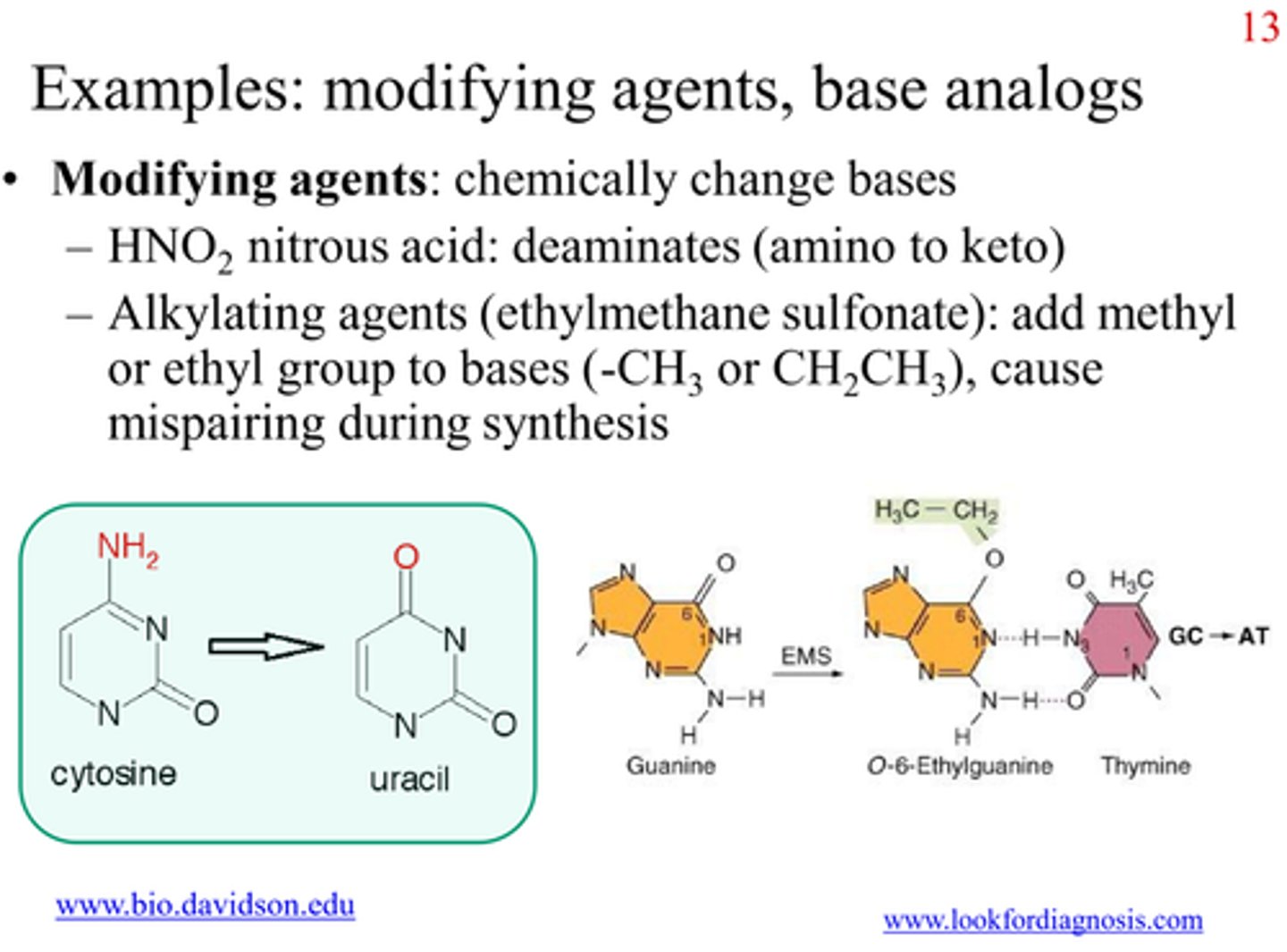
Hydroxylamine (NH2OH)
- hydroxylating mutant that reacts specifically with cytosine
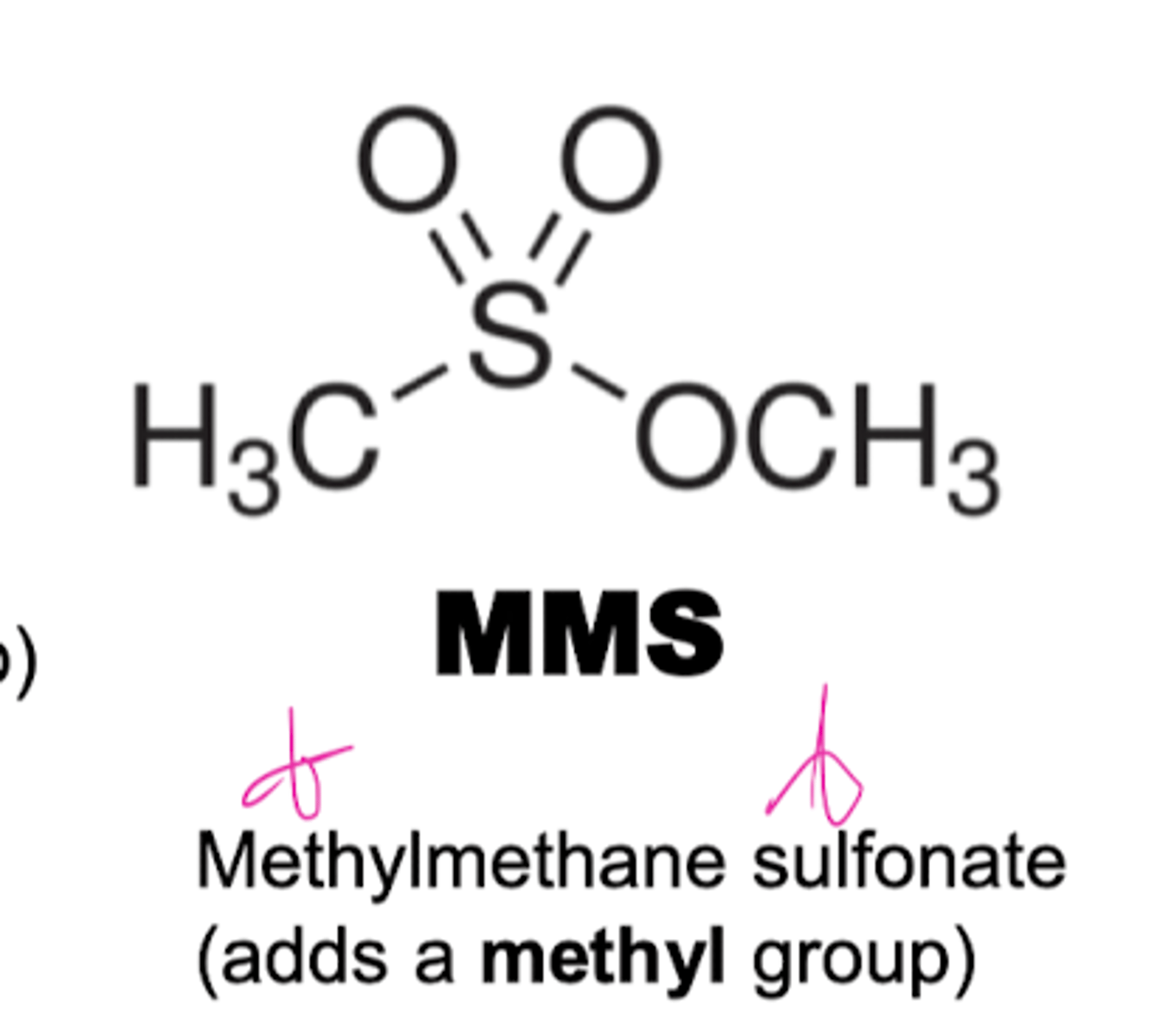
Methylmethane sulfonate (MMS) alkylating agent
ntroduces alkyl groups onto bases.
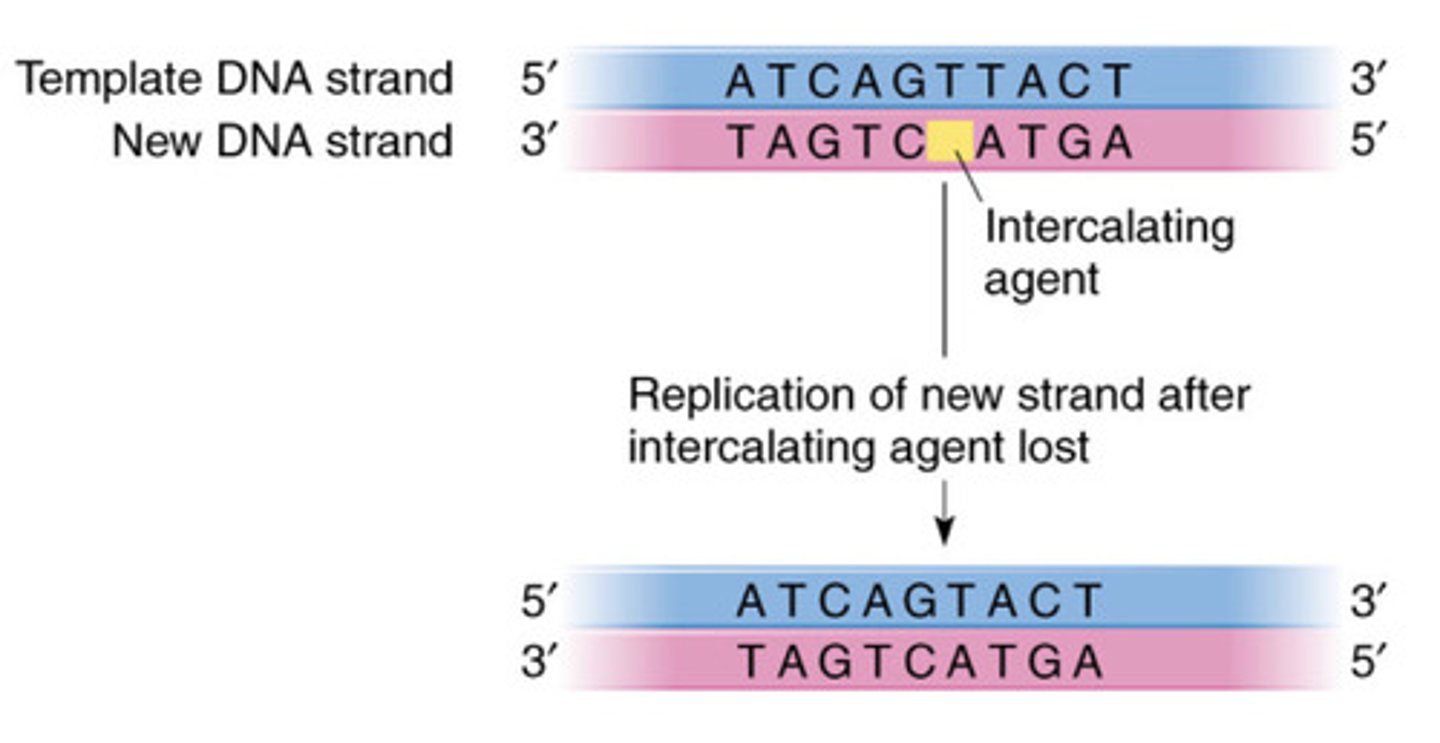
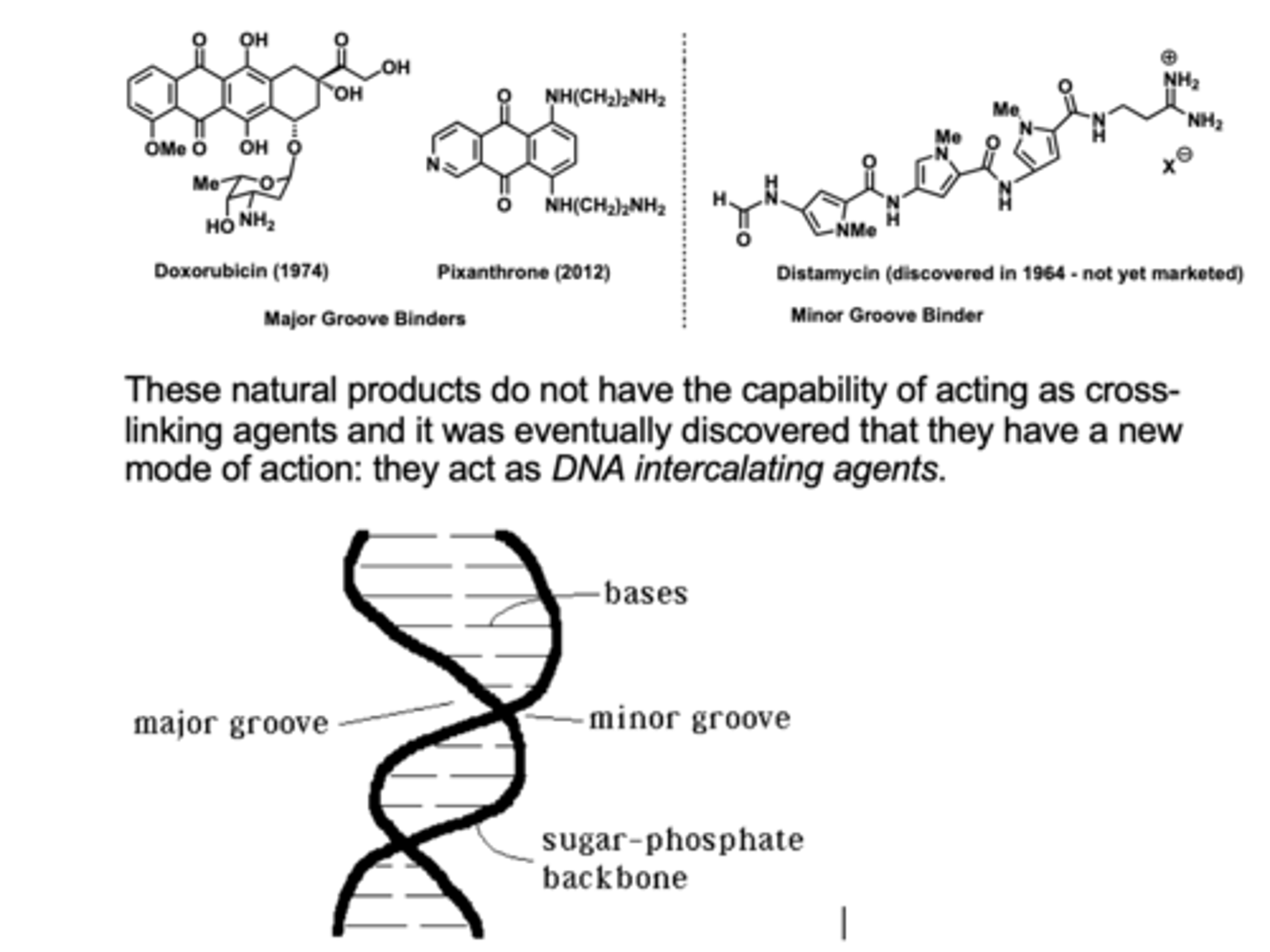
3.Intercalating Agents
•Chemicals which insert themselves into the DNA double helix
•Can lead to addition or deletion of bases
- Examples: ethidium bromide,
anti-cancer drugs, e.g. doxorubicin, bleomycin
Intercalating Agents - Addition
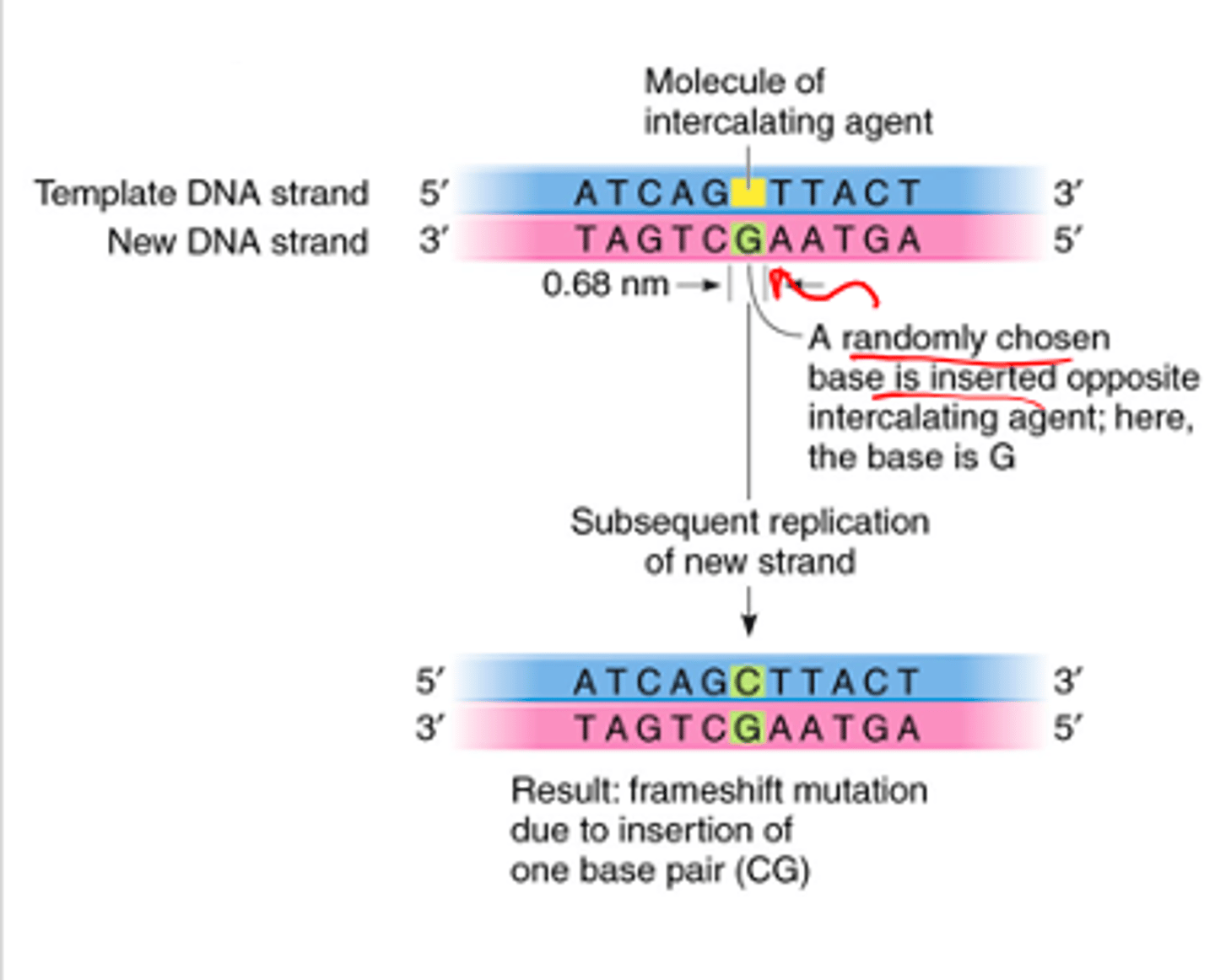
Intercalating Agents - Deletion