12. Site Specific Recombinations
1/55
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
56 Terms
site specific recombination occurs between sequences with a
limited stretch of similarity; involves specific sites (recombination sites)
regions with similar sequence will “flank” DNA
that can be moved or rearranged
three types/functions of site specific recombination
inversion
insertion
deletions
inversion e.g
expression of alternative genes - inversion of DNA sequence/gene by DNA invertases
Insertion e.g
infection - insertion of bacteriophages in the bacterial genome by DNA integrase
Deletions e.g
reversal of insertion, mediation of programmed DNA rearrangement during embryogenesis
components of the site-directed recombination are
rearrangement enzymes
recombination site
rearrangement enzymes - recombinases (in some cases accessory proteins are required0
site specific recombinases cleave and rejoin DNA using a covalent protein-DNA intermediate
a) site specific endonuclease activity (cleavage)
b) DNA ligase activity (rejoining)
Recombination site (specific sequence) places where DNA exchange will occur
c) recombinase recognizes sequence
d) non-palindromic (asymmetric) sequences
recombination sites in SAME ORIENTATION, TWO different DNA molecules
insertion (integration; lambda integrase)
recombinatino sites in SAME orientation, same DNA moolecule
deletion (excision)
Recombination sites in INVERTED orientation, same DNA moelcule
Inversion* (DNA invertase)
DNA deletions and insertions used ro recycle genes
recombination sites are recognized by specific recombinase
recombination sites are flanking the gene (e.g antibiotic resistance gene) in direct orientation and on the same DNA molecule
steps for DNA deletions and insertions
recombination sites align next to each other
recombinase resolves the structure and cuts out the insert gene
both the insertion of the lambda phage DNA into bacterial genome and its deletion from it are accomplished by
site-specific recombination event, catalyzed by the lambda integrase enzymel insertion invovles some bacterial enzyme as well
when the lambda DNA enters the cell, the ends join to form a circular DNA molecule, it can go in two pathways
prophase pathway
insert its DNA into bacterial genome and enter a latent prophage state
lytic pathway
lambda bacteriophage can multiple in in E.coli, cell lysis releases a large number of new viruses and destroys cell
prophage can exit the host chromosome and shift to
lytic growth (induction)
DNA invertases
recombination sites are repeated sequences but in inverted orientation on the same molecule DNA molecule
recombination sites flank the DNA sequence which is to be inverted
STEPs for DNA inversion
recombination sites align next to each other
cross-over and resolution by recombinase → DNA sequence is inverted
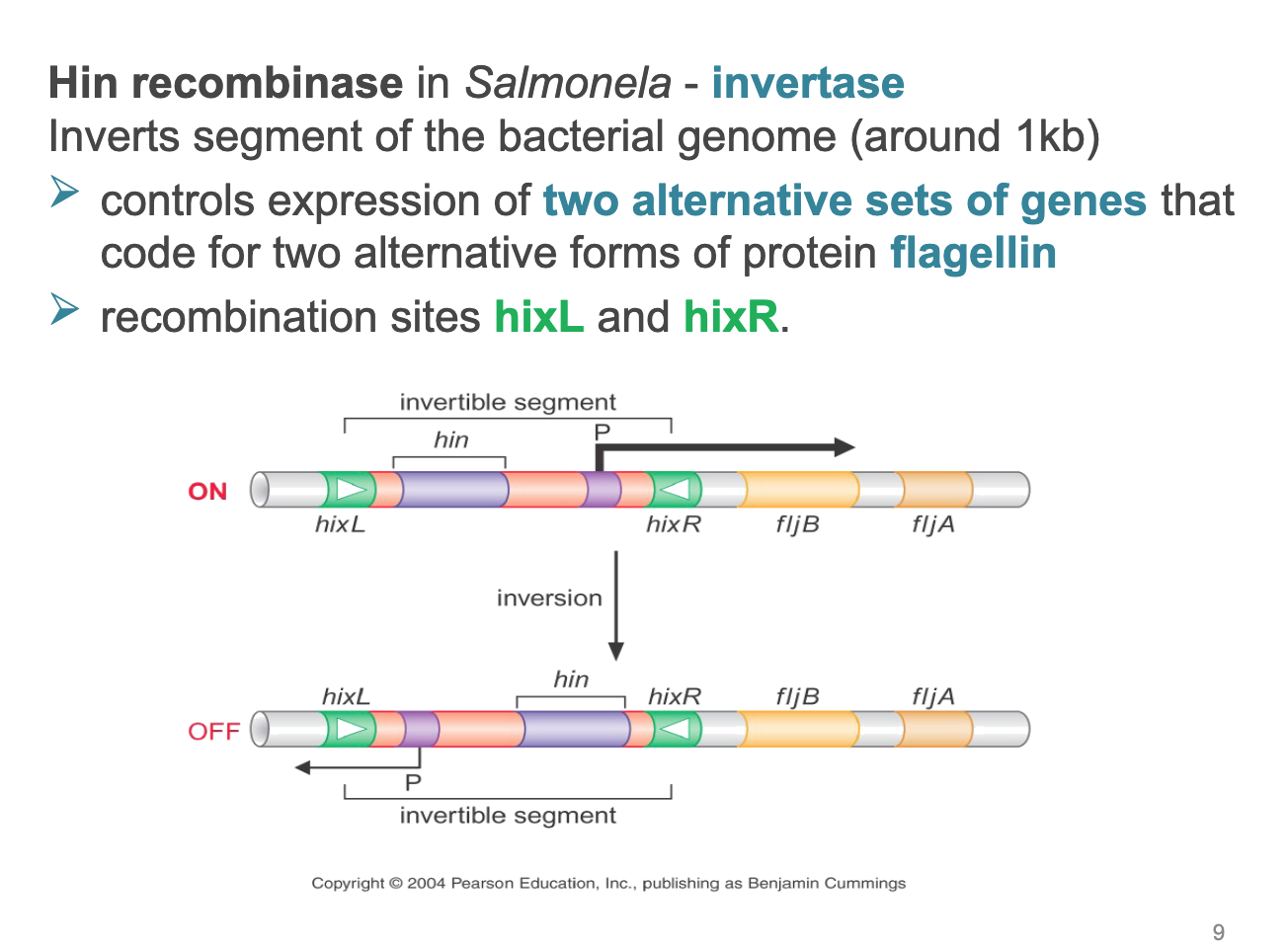
Hin recombinase in Salmonela - invertase inverts segment of the bacterial genome (around 1kb)
controls expression of two alternative sets of genes that code for two alternative forms of protein flagellin
recombination sites hixL and hixR
nonhomologous recombination
transposable (mobile, transposon) elements insert into DNA that has no sequence homology with the transposon
two sites in transposition
donor site
contains a transposable element (transposon)
target site
usually random, however, there are hot spots; preferred sequences that are targeted
autonomous
encode all the enzymes necessary to move
nonautonomous
have no coding capacity (their mobility depends on the enzymatic machinery of their autonomous “relatives”)
non-replicative transposition
“cut and paste”
both strands of original transposon DNA move together from 1 place to another without replicating
replicative transposition
copy and paste
involves DNA replication phase → 1 copy of a transposon remains at original site
new copy inserts at the new site
transposons can cause genetic changes
(horizontal transfer - contributions to the evolution of genomes)
transposons insert
into genes/coding sequences
regulatory sequences (induce change in gene expression)
transposons can also form
chromosomal rearrangements and relocate genes
insertion of transposon in protein-coding region
disrupts the protein sequence
transposon element insertions represent the major source of
spontaneous mutations in drosophila melanogaster
ultrabithorax mutant
double thorax/wing mutation is due to insertion of Doc transposable element into the first exon of the Ubx gene
in humans, transposon elements
Haemophilia A: insertions of L1 element into exon 14 of the factor VIII gene in two of 240 unrelated patients with haemophilia A
Primary breast cancer: insertion of Alu in BRCA2 gene
however, possible positive effects of transposition - domestication of genes carried by transposons
systems with transposon- derived genes thru domestication
initiation of meiotic recombination (possible role remodelling)
regulation apoptosis
control of cell cycle
defense from transposon invasion
regulation of transcription
DNA-only transposons
short inverted repeats at each end
retroviral-like retrotransposons
directly repeated long terminal repeats (LTRs) at each end
nonretroviral retrotransposons
poly A at 3’ end of RNA transcript; 5’ end is often truncated
LTR
long terminal repeats exist in retroviruses; substitute “LTR” with retroviral-like
DNA-only Transposons: Insertion Sequence (IS) elements
simplest type of transposable element found in bacterial genomes and plasmids
encode only genes for mobilization and insertion (host replication machinery used for replication)
transposon size from 768 bp to 5 kb
end of all IS elements show Inverted Terminal Repeats (ITRs)
Retroviruses:
exist as a ss RNA genome packed into capsid, together with reverse transcriptase (protein) and the dsDNA form has terminal repeat sequences (LTR = direct long terminal repeats)
retroviral like
LTRs
RNA (intermediary)
codes for enzymes
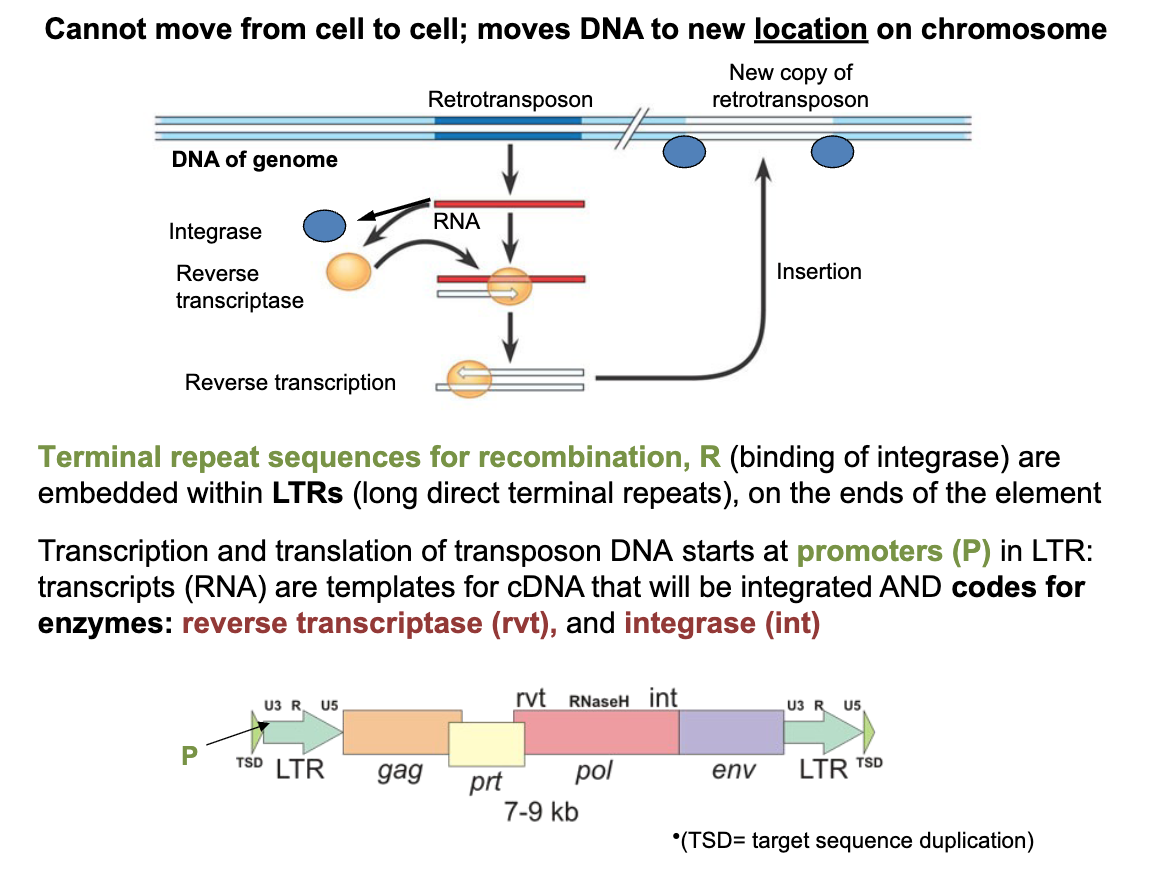
Retroviral-like retrotransposons - cannot move from cell to cell; moves DNA to new location on chromosome
Terminal repeat sequences for recombination, R (binding of integrase) are embedded within LTRs (long direct terminal repeats) on the ends of the element
in retroviral-like retrotransposons: transcription and translation of transposon DNA starts at
promoters (P) in LTR: transcripts (RNA) are templates for cDNA that will be integrated AND codes for enzymesL reverse transcriptase (rvt) and integrase (int)
LINEs (long-interspersed sequences)
up to 6.5 kb sequences, repeated 50,000-100,000X (~5% of genome)
presumable transcribed from internal (downstream) RNAP II promoter (do not have LTRs)
majority LINEs encode two ORFs whicha re transcribed as a
bicistronic mRNA composed of ORF1 (RNA binding protein, etc.) and ORF2 (endonuclease and reverse transcriptase activities)
code for proteins necessary to reverse transcribe and integrate SINEs as well
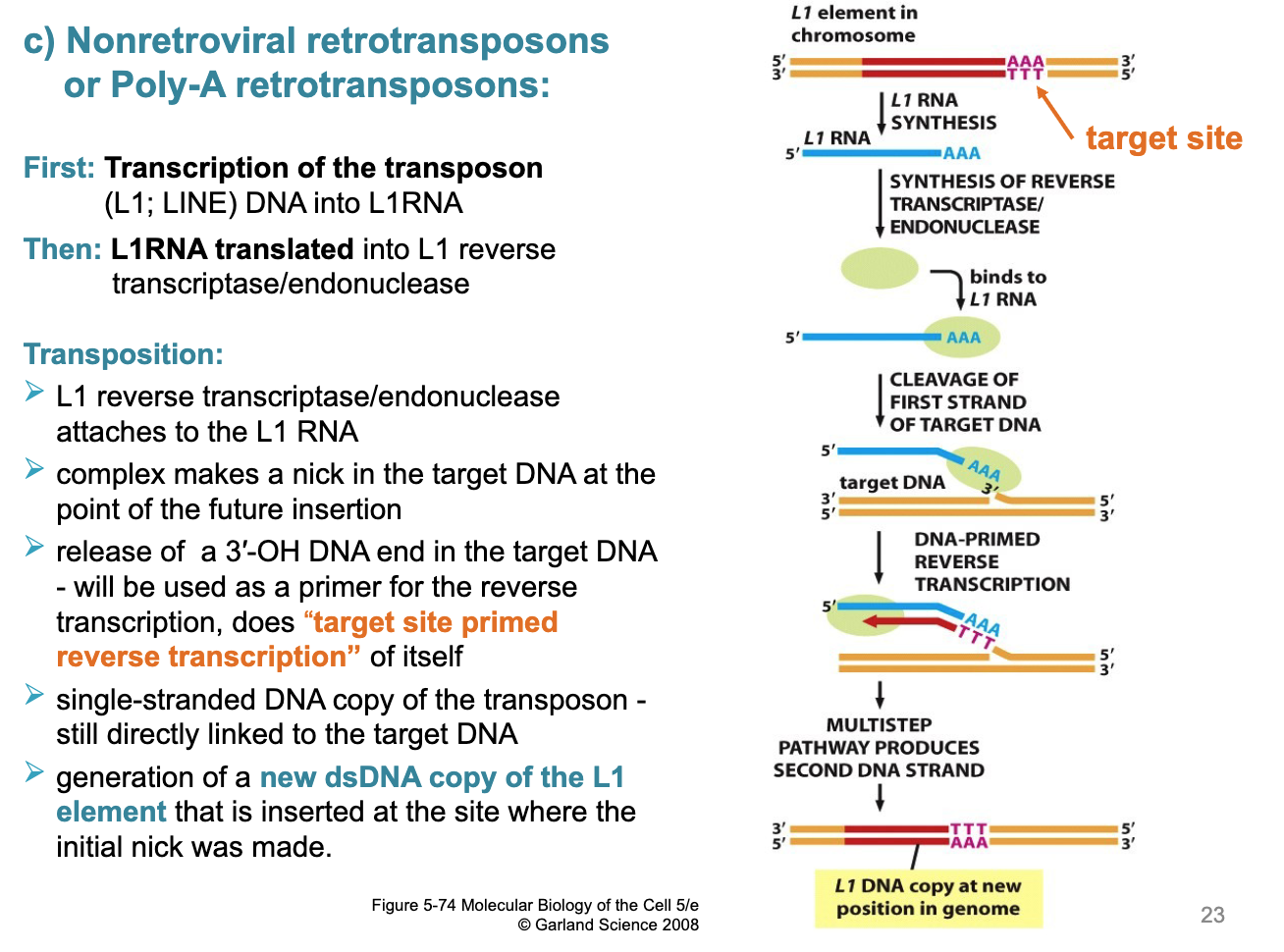
first: transcription of the transposon (L1: LINE) DNA into L1RNA then
L1RNA translated into L1 reverse transcriptase/endonuclease
Transposition in nonretroviral retrotransposons or Poly-A retrotransposons
L1 reverse transcriptase/endonuclease attaches to teh L1 RNA
complex makes a nick in the target DNA at the point of the future insertion
release of a 3’OH DNA end int he target DNA will be used as a primer for the reverse transcription, does “target site primed reverse transcription” of itself
single-stranded DNA copy of the transposon still directly linked to the target DNA
generation of a new dsDNA copy of the L1 element that is inserted at teh site where the initial nick was made
SINEs (short-interspersed sequences)
~ 300 bp long, repeated 300,000 - 500,000X
flanked by 7-20 bp direct repeats
do not code for proteins so they cannot transpose by themselves
smoe are transcribed into RNA intermediate (contain poly-A tails for priming reverse-transcription, but reverse transcriptase/endonuclease machinery used is from LINEs)
evolutionary origin of SINEs
tRNA gene or other small RNA genes
humane genome: Active SINE element is called Alu
during the cellular heat shock response, human Alu RNA blocks transcription by binding to RNAP II; it enters into transcriptional machinery complexes at promoters in human cells in vitro - acts as a transacting, promoter specific, transcriptional repressor
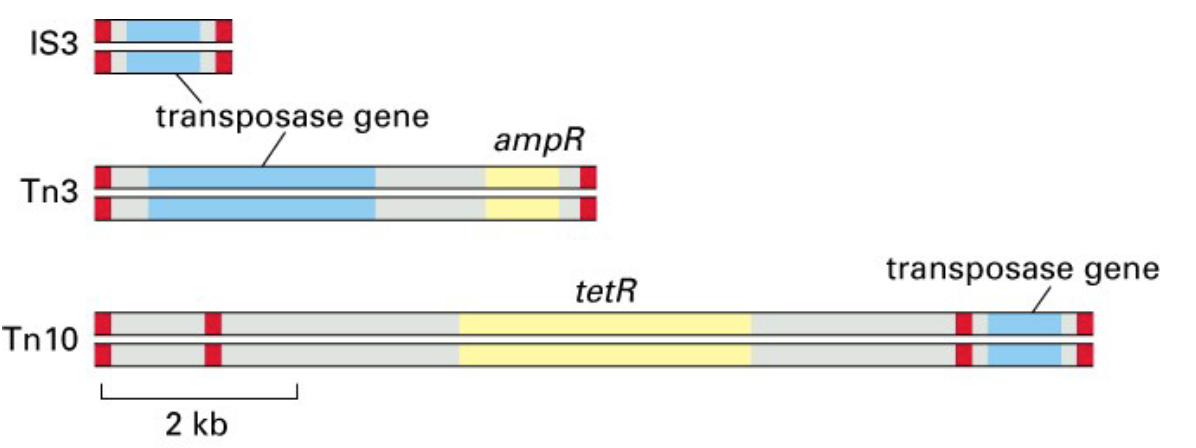
transposon elements found in bacteria:
transposase gene: encoding enzymes for DNA breakage and joining
red segment: DNA sequences as a recognition sites for enzymes
yellow segments: antibiotic resistance genes
barbara mcClintock
nobel prize in physiology and medicine 1983: for her discovery of mobile genetic elements
studied transposable elements in corn
McKlintock’s discvery
muation and its reversion result from Ds (dissociation) element
transposes into the C gene
mutates pigment gene (yellow)
transposes out again; color reverted to wild type (purple)
Ds cannot transpose by itself, must be a
non-autonomous transposon, must have help from an autonomous transposon, Ac (for activator)
Ac carries transposase, Ds is an AC element with most of its middle removed
DS has only
pair of inverted terminal repeats (insertion into C)
adjacent short sequences that Ac transposase can recognize
spotting in the maize kernels
multiple reversions of an unstable mutation in the C locus (responsible for purple kernel colour)