BIS2A SS1 Week 4
1/61
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
62 Terms
3 ways that cells deal with toxic oxygen buildup
1) move away from oxygen
2) detoxify oxygen (using catalases, etc.)
3) detoxify and use oxygen (aerobic respiration)
what allowed eukarya to expand processes from bacteria and archaea?
increased ATP production due to aerobic respiration
what must a cell do?
reproduce (“viable” cell)
some organisms are metabolically active but do not grow (stationary phase)
what is the purpose of the cell membrane
to differentiate between the inside and the outside of the cell (a cell wall counts as the outside)
advantages to being small
1) can move things around cell via diffusion
2) less energy usage
3) lower surface area to volume ratio
4) size — can get away from predators
disadvantages to being small
1) vulnerable to predation
2) limited by size — can’t move to a new environment with nutrients and less storage room
3) more competition
do organisms want to be smaller or larger?
larger — less likely to be prey, can move around to get nutrients
diffusion is a problem though
eukaryotes vs prokaryotes
eukarya have membrane bound DNA (nuclei)
prokarya do not
are eukarya more closely related to archaea or bacteria? what process in the cell gives this away?
archaea and eukarya are more closely related to each other vs to bacteria
they have similar information storage and transmission processes
why are processes conserved? where can the information for these processes be found?
processes like SLP are conserved because they’re useful; any unnecessary information is removed
ex. parasites have small genomes because they don’t need a lot of information and can make host do their processes
capsule
acts like the garage of the cell; can store stuff like sugars, etc. (between cell wall and cell membrane)
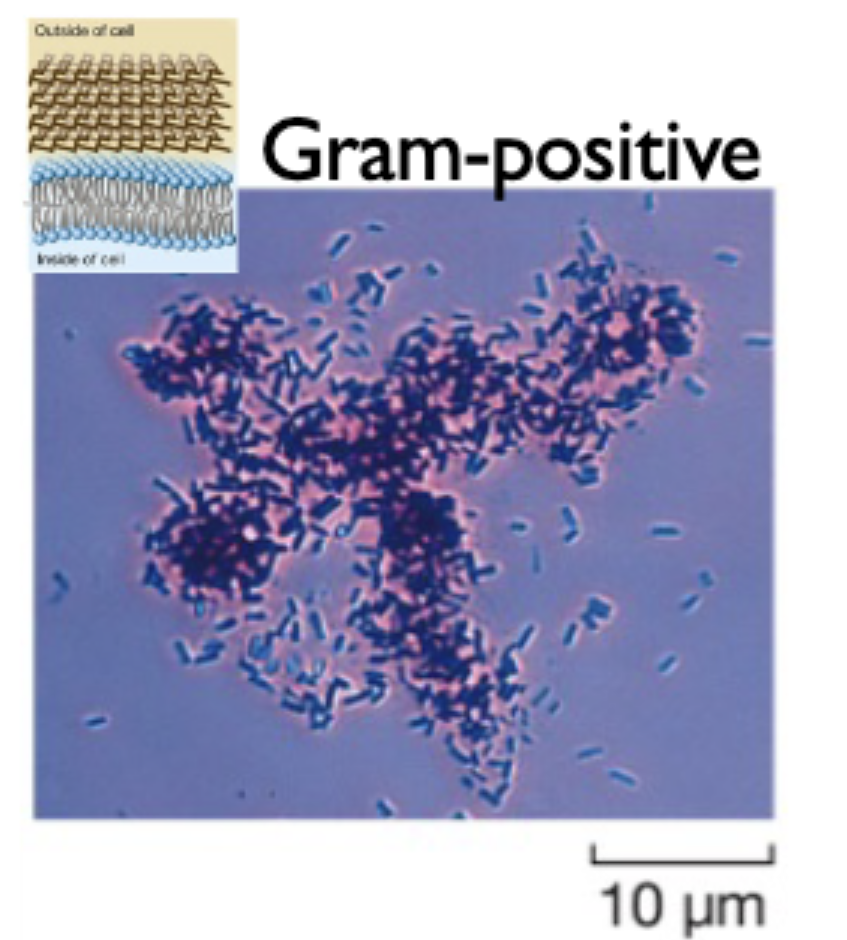
gram positive bacteria
have a thick peptidoglycan cell wall that stains purple
disaccharaides in cell wall are linked by peptides in a 3D manner
sensitive to penicillin/ampicillin that cleave peptidoglycan
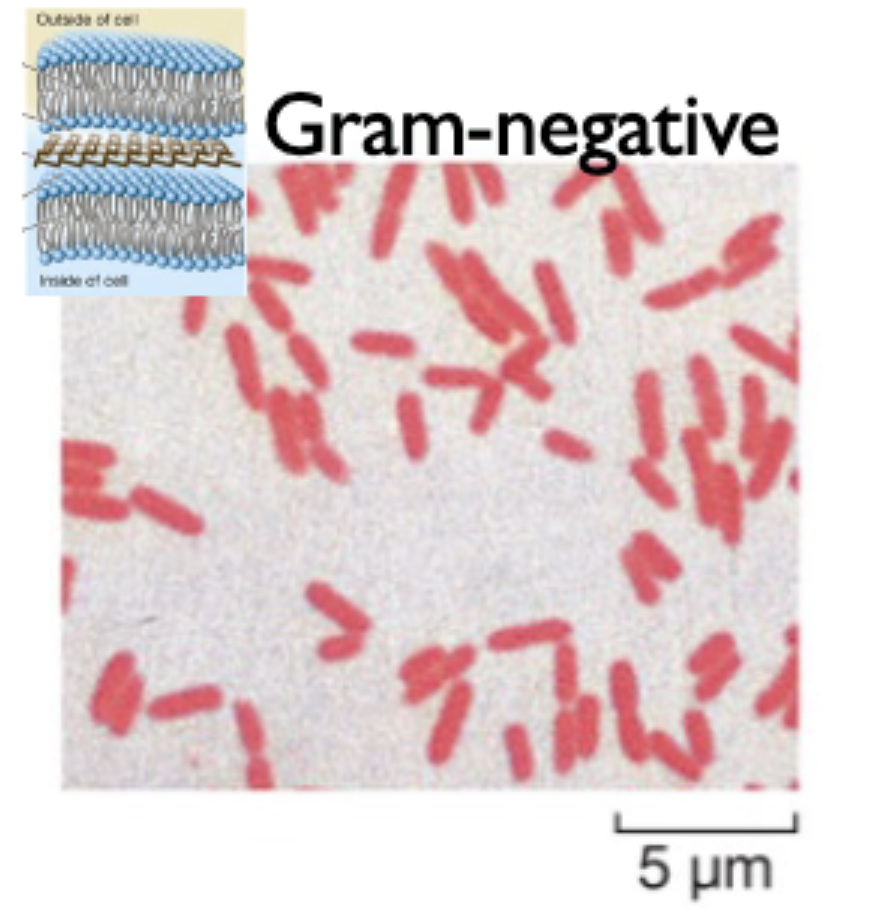
gram negative bacteria
have a thinner peptidoglycan cell wall in between two lipid bilayers; stains red
outer membrane made of LPS not phospholipids
periplasm (area between cell wall and membranes) is where activity occurs
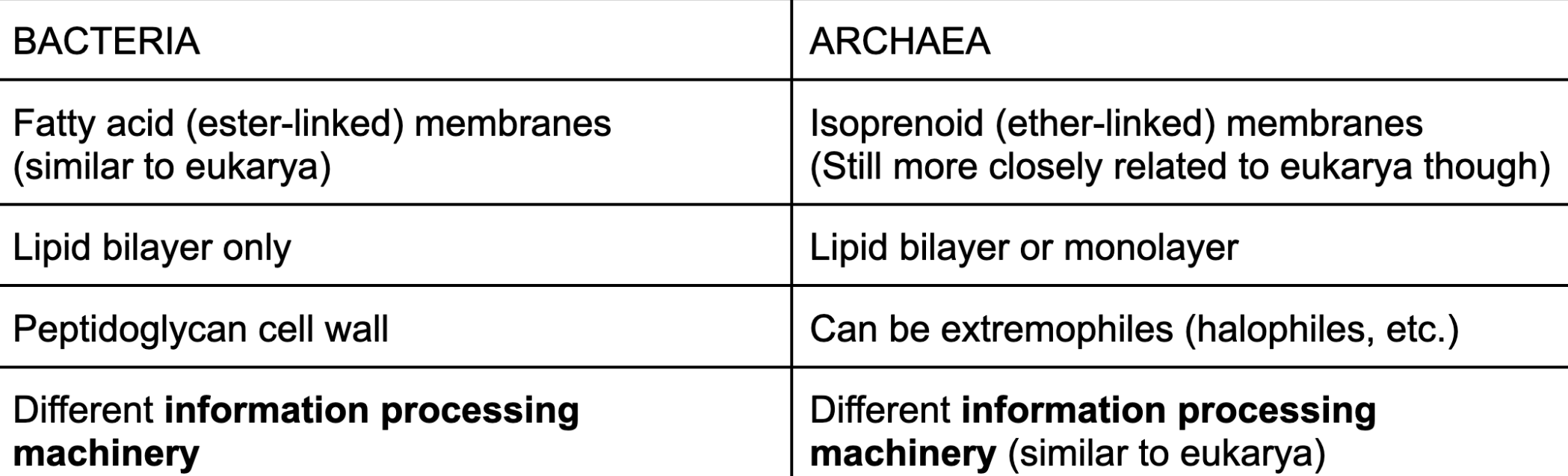
bacteria vs archaea
similarities: similar metabolism (generate energy, degrade and build material)
differences in picture
why is increased surface area important for cells?
electron transport, which produces energy, is limited to cell membranes
more surface area increases ATP production
cells want to increase surface area without actually getting larger
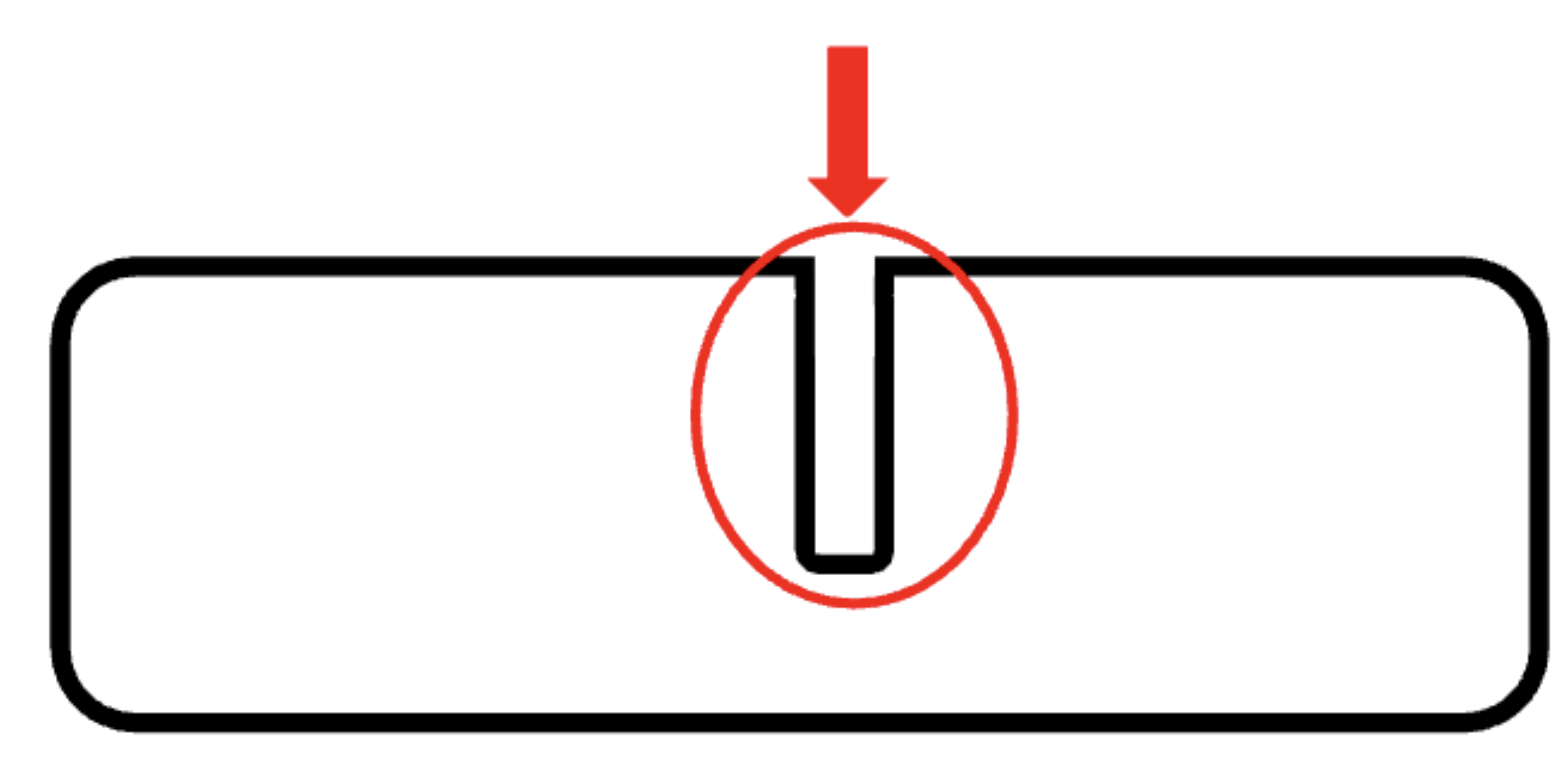
what is the mechanism to increase surface area while keeping volume low?
invaginations (create a folded membrane)
1 mutation in e. coli can cause this
which organisms take advantage of invaginations
photosynthetic bacteria (increase surface area for PS, ETC, etc.)
rapidly respiring bacteria (anaerobic and aerobic)
what is the benefit of getting large?
compartmentalization
benefits of compartmentalization
1) solve diffusion problem for pathways with many steps
2) sequesters “bad” reactions from cell (ex. lysosomes require a low pH environment)
3) greater regulatory control/minimizes cross-talk
origin of eukaryotic organelles
endosymbionts like mitochondria (first one) and chloroplasts live inside eukaryotic cell, becoming in sync with host and eventually losing abilities to make compounds/DNA for that (dependent on host)
what is DNA used for? what are some key features of DNA?
DNA is used for information storage
made of two antiparallel complementary strands (one runs 5’ to 3’ and vice versa for other strand)
4 types of information storage in viruses
double stranded and single stranded DNA
positive RNA virus — genome is like mRNA
negative RNA virus — made in to + RNA then mRNA
DNA-dependent DNA polymerase
reads DNA strands to make its complement
reverse transcriptase
converts RNA to DNA
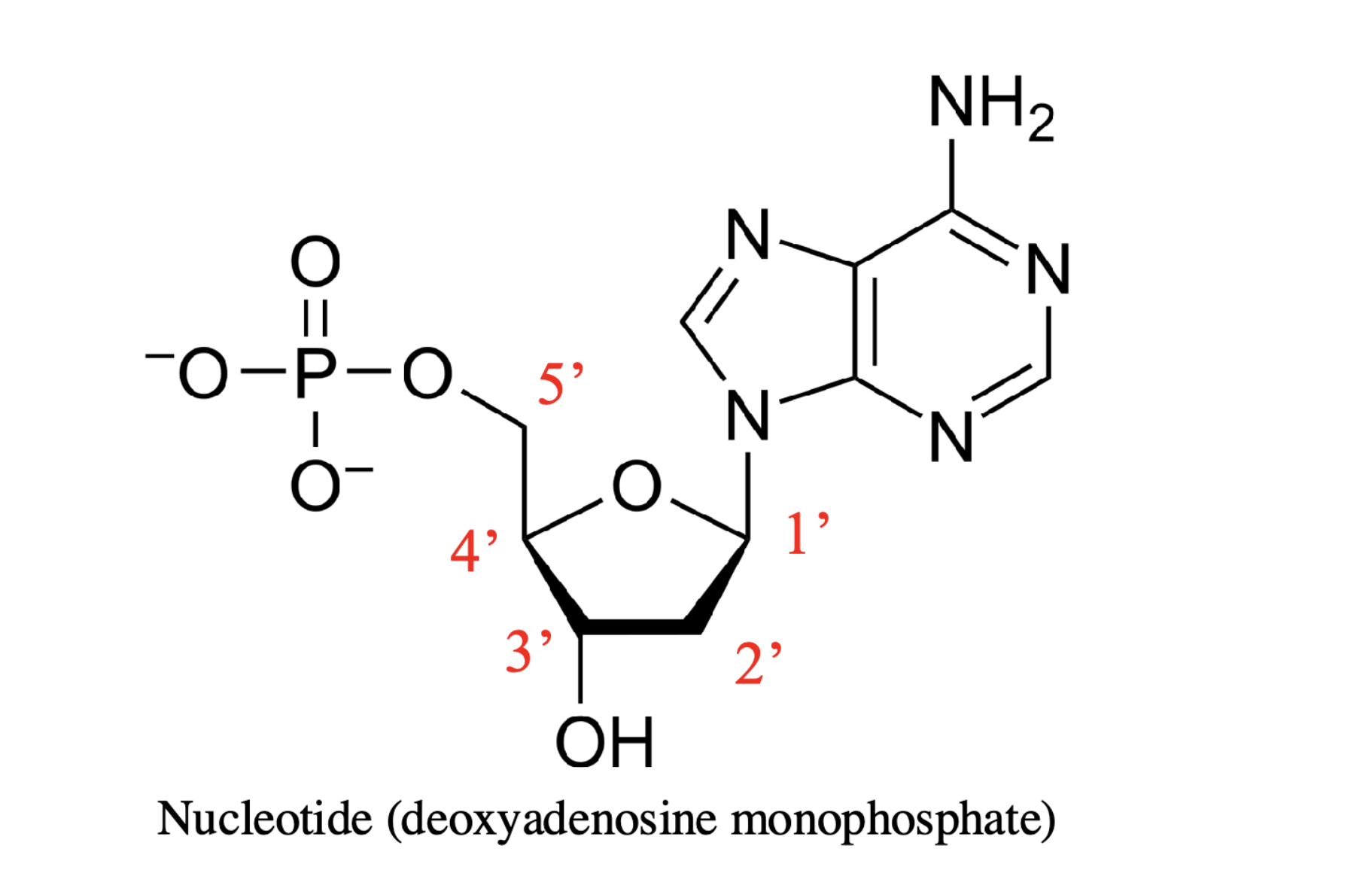
key features of nucleotide
1’ - nitrogenous base
2’ - hydroxyl or hydrogen (determines ribose vs deoxyribose)
3’ - hydroxyl
5’ - phosphate groups
purines
double nitrogen ring — adenine and guanine
remember by “pure as gold”
pyrimidines
single nitrogen ring — cytosine, thymine, uracil
how many hydrogen bonds between adenine and thymine/uracil? between cytosine and guanine?
2 between A/T+U; 3 between CG
these hydrogen bonding patterns and order of nitrogenous bases store info
key features of DNA
naturally helical
negatively charged (proteins that bind have a positive charge)
contains a major and minor groove (due to antiparallel pattern of DNA)
flexible overall, shorter segments are rigid
what makes the DNA structure strong
the many stacking H bonds (external H bond donor/acceptor contribute to DNA solubility
major reason cell does not like single stranded DNA
it is a sign of DNA damage and ends, which cells don’t like
can also interfere with normal base pairing of DNA
how do cells protect single stranded DNA from being attacked
single stranded binding proteins
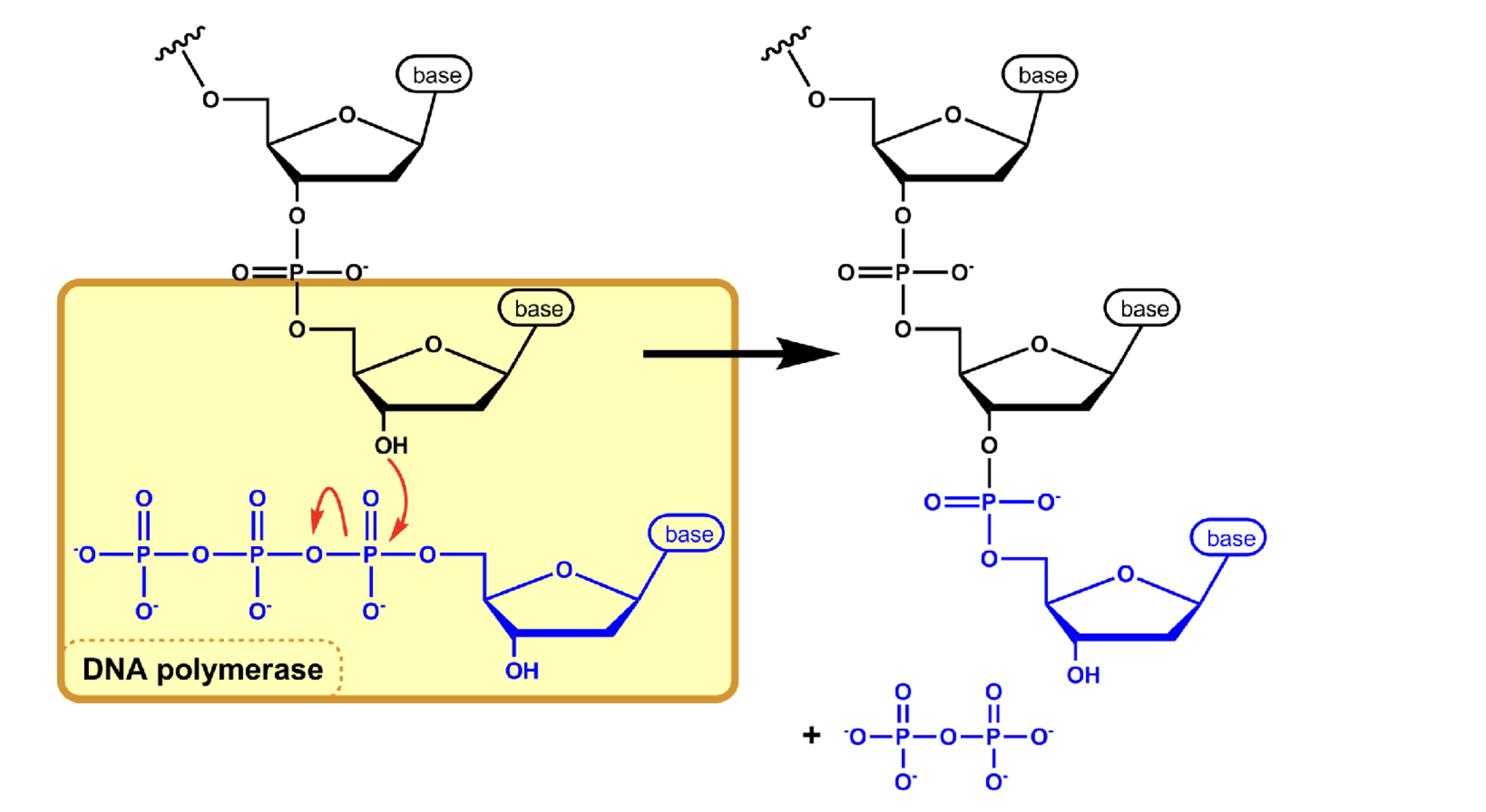
phosphodiester bond
polymerization reaction between the phosphate (5’) and the hydroxyl (3’) to add incoming nucleotides (dNTPs) to a growing chain — polar and created between O and P
only added to 3’ end
where does DNA replication begin?
at the origin of replication (ORI-C in bac/archaea) where a replication bubble (with two replication fork that extends in opposite way) is formed
replication begins in the middle of the bubble
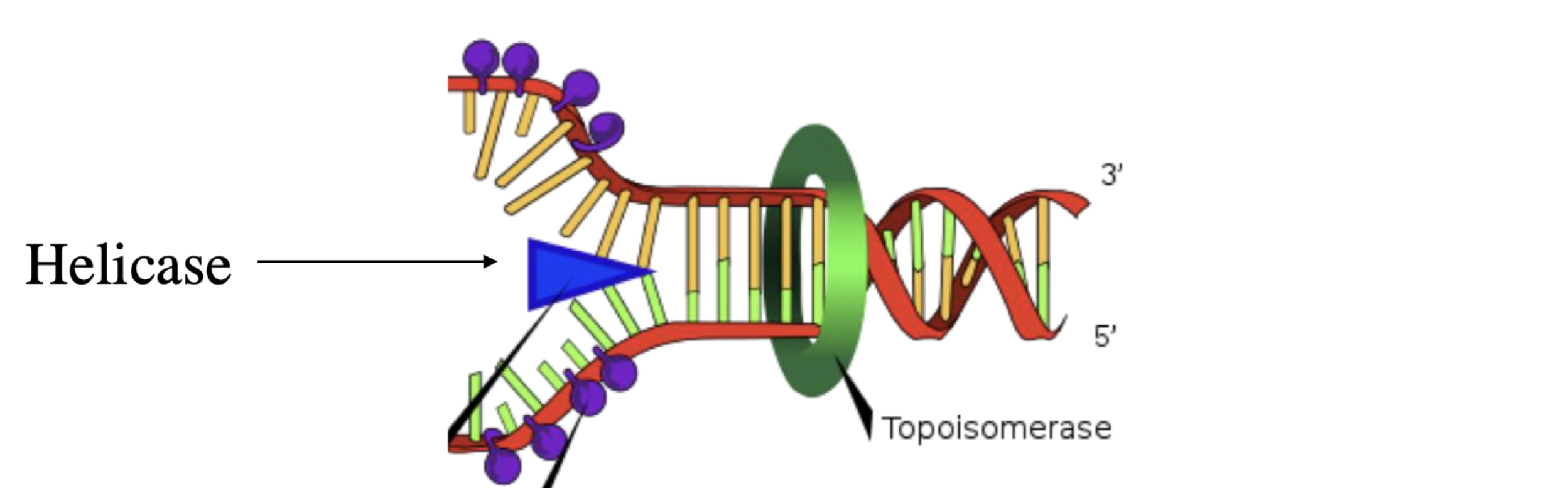
what is the key enzyme in the initiation of replication?
helicase, DNA-A, single stranded binding protein (SSBPs)
DNA-A
binds to ori-c to open the double helix, consuming itself (stoichiometric enzyme)
single strand binding proteins
stabilize the open double helix
helicase
“unwinds” the DNA double helix by breaking the hydrogen bonds between the nitrogenous bases
also winds up the DNA (positive supercoiling) and unwinds DNA (negative supercoiling) which costs ATP
elongation of replication
synthesizing the daughter strands
in which way is the parent strand read? the daughter strand synthesizes?
parent strand read from 3’ to 5’
daughter strand synthesized from 5’ to 3’
key enzymes in elongation
RNA primase, DNA polymerase III, DNA polymerase I, ligase, SSBPs
RNA primase
creates a temporary RNA primer that provides a free 3’ hydroxyl group for DNA polymerase III to begin synthesis
DNA polymerase III
adds complementary DNA nucleotides and elongates daughter strand
is processive (doesn’t want to be unbound from DNA)
DNA polymerase I
replaces the RNA primers with DNA nucleotides
ligase
joins the backbones/seals nicks of the okazaki fragments together in lagging strand — connects the 5’ phosphate and 3’ hydroxyl
leading strand
the strand that is continuously synthesizes from the 5’ to 3’ direction; synthesizes towards the replication fork
lagging strand
the strand that is discontinuously synthesized in the 5’ to 3’ direction; requires many primers, DNA fragments are called okazaki fragments
synthesizes away from the replication fork
requires many SSBPs as it is synthesized in parts
what bond is created by ligase to link the okazaki fragments together?
phosphodiester bonds
in bacteria, does the replication fork move or the DNA?
DNA
termination of replication
when replication, there is a 3’ overhang present in lagging strands where the RNA primer was removed and DNA polymerase I cannot add bases (no free 3’ OH)
terminus
opposite of oriC where replication bubble meets, where replication stops and the last set of ends are joined
terminase
enzyme that separates the two replicated circular chromosomes
ter site
one-way end site to prevent the enzymes from continuing replication (DNA pol III can go through but stops at other site)
telomeres
non coding regions at the end of chromosomes that are stretches of junk DNA
stabilizes DNA structure and prevents DNA shortening
telomerase
lengthens the telomerase by recognizing the repeated patterns of junk DNA and lays down more nucleotides to lengthen telomere using an RNA template to provide a free 3’ OH for DNA polymerase
is a ribozyme
how do telomerases know what to add?
contains an RNA template to create the repeated sequence at the end
how are telomeres linked to aging?
as telomeres get shorter, we age
what are two requirement for replication?
fast and accurate
if too fast, leads to mutations, but we need some mistakes for evolution
why are mutations in DNA beneficial?
increases genetic diversity in the population, which increases its overall health
why can e.coli/fast growing bacteria replicate faster than their DNA can be replicated?
they initiate replication in intervals equal to their division time (ex. 20 mins for e.coli)
results in multiple replication bubbles at the same time, leading to overlapping cell cycles in fast growing bacteria
how many polymerases are active at once in e. coli/prokaryotes?
4 polymerases (bidirectional from a single origin — 2 strands, 2 directions)
how do eukaryotes replicate so fast?
contain many origins of replication, which costs a lot of energy (ATP) bc of many helicases