New Material Lecture 1/5: Mutation and DNA Repair
1/42
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
43 Terms
what are mutations
changes in nuc sequences that get passed on to daughter cells
new alleles are created by muts of an existing allele
muts that occur in germ cells can be passed on to offspring
why are mutations important for life
evolution/adaptation/genetic diversity
why are there mechanisms to fix muts
lethal muts
too many muts ruin the integrity of a species
no maintenance of key genetic/allele combos
eg if trait promotes fitness, parents must be able to mostly pass that onto the offspring
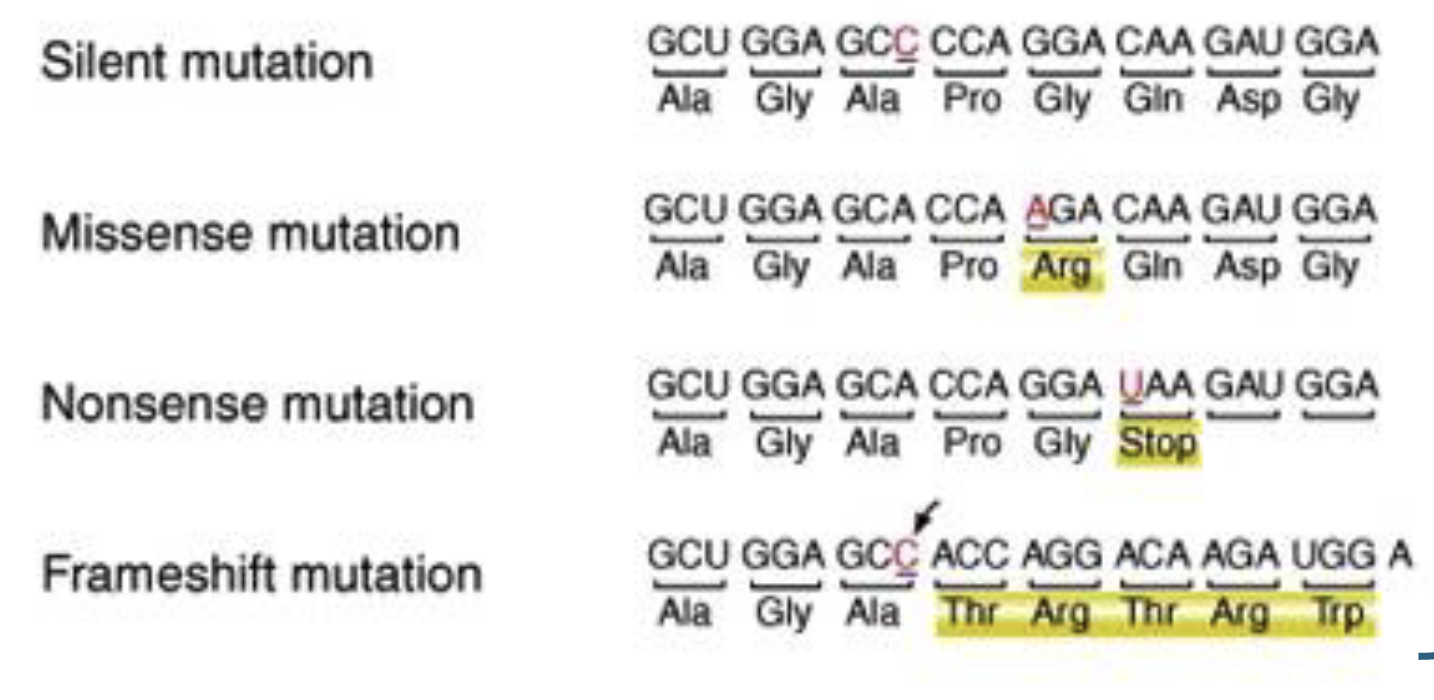
what are substitution muts
replacement of one base by another base
what are purines and pyrimidines
Purines (adenine and guanine) are two-carbon nitrogen ring bases
pyrimidines (cytosine and thymine) are one-carbon nitrogen ring bases
what are transition and transversion muts examples of? explain what each is
The 2 types of substitution muts
transition → purine replaced by purine or pyaimidine by pyrimidine
transversion → purine replaced by pyrimidine or vice versa
what are deletions
one or more bps are lost from dna
what are insertions
one or more bps are added to dna
how many bps are deleted/inserted in those types of muts
can involve one or few to hundreds/thousands of bases
they’re not ALL frameshifts if multiple of 3
what is a point mut
umbrella term → any mut that is only affecting a single nucleotide (sub, insertion, deletion)
what are frameshift muts
when deletions or insertions change reading frame of gene
causes AAs incorporated during translation downstream of region where deletion/insertion occurred
can introduce early stop codons
what are inversion muts? why do they occur
when a segment of dna is reversed (inserted backwards)
occurs when two dsDNA (double-stranded) breaks occur and excised dna is added back in the incorrect orientation
do all muts lead to diff phenotypes
no → can have silent muts
a lot of dna does not code for AAs (eg introns)
coding is redundant so sometimes changes in bps may j result in same AA produced
t/f: spontaneous muts occur at a fast rate
f → they occur at a very slow rate

are muts heritable in euk organisms
only muts in germ cells are
muts in somatic cells are passed to daughter cells in that individual
are muts heritable in proks
yes → always j make copies of themselves so all muts are heritable
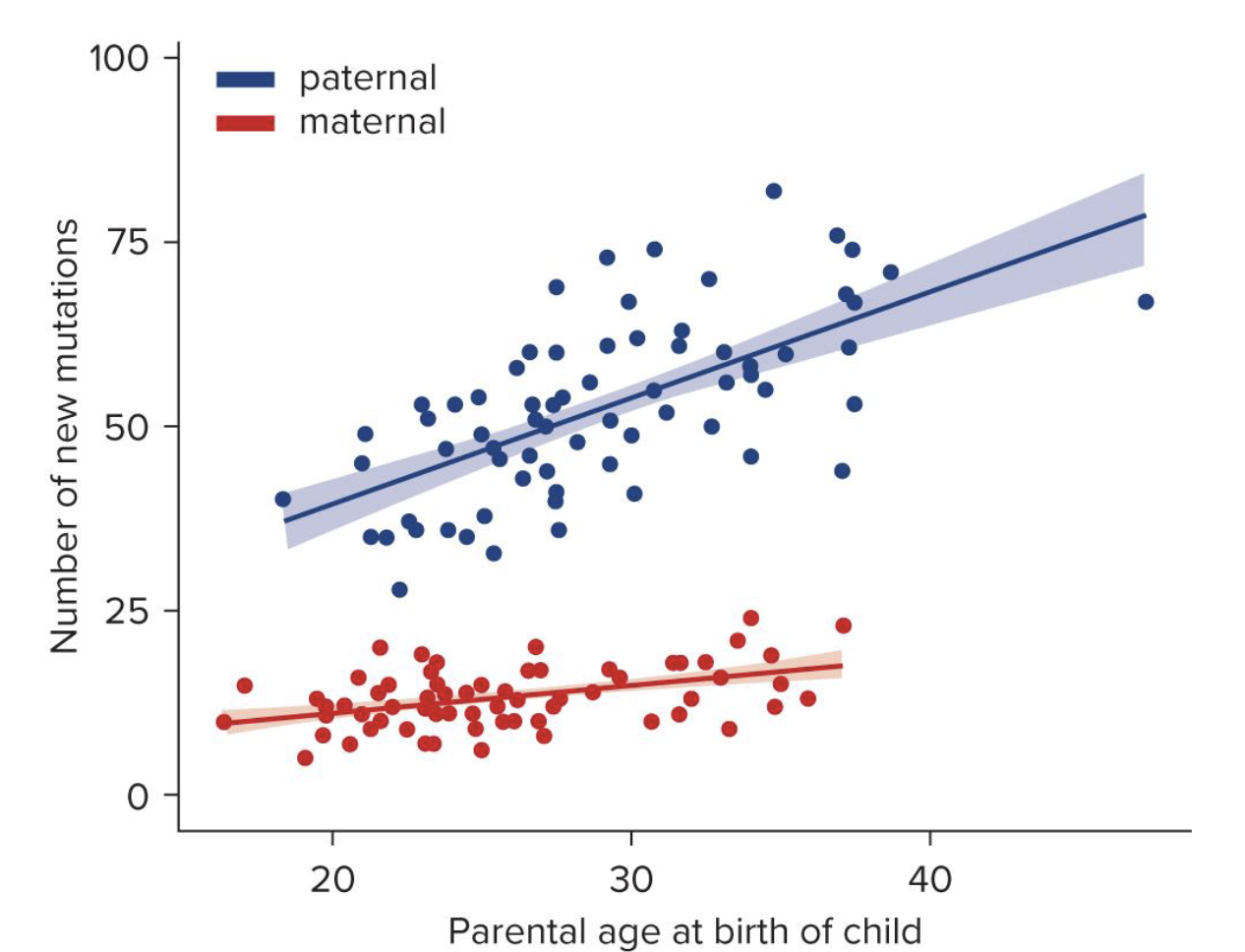
t/f: sperm have more muts than eggs
explain why or why not
true → long-term arrest of meiosis in egg cell precursors during diplotene 1 inc the occurence of non-disjunctiuon during oogenesis compared to spermatogenesis
BUT spermatogenesis is ongoing → occurs en mass so inc chance of muts and passing on those muts to offspring (bc germ cells)
t/f: muts in maternal germ cells greatly inc w age and paternal germ cell muts are relatively constant through life
false → opposite
explain the experiment that showed us if muts occur randomly or bc of environmental stress
let ecoli cultures grow for specified aount of time
plated equal portions of each culture w basteriophage T1
if muts occured bc of environment, would be around equal proportions of each culture that are resistant (bc muts would ONLY occur after phages were introduced)
if random, they varying amounts
found they were random
muts occurring early in colony development resulted in many resistant colonies when plated; those occurring later resulted in few resistant colonies
how did replica plating verify that bacterial resistance is result of preexisting muts
agar plate contained colonies of type of bacteria killed by penicillin
created “stamps” of those cells and transferred it to an agar plate containing penicillin
most cells would die but cells w resistance would form new colonies
position of resistant colonies would tell if colonies on OG plate ALREADY had resistance if resistance predated interaction w pen
if already had resistance before pen, all colonies on agar plates would be immune in same areas
if developed resistance after pen, would be immune in diff areas
found colonies on agar plates were all resistant in same areas → hence had resistance before pen → hence muts are random
t/f: selective pressures cause mutations
false
what are depurination and deamination? explain them
they are processes that lead to muts
Depurination: hydrolysis (removal) of purine bases
1000/hour in every cell
random bp introduced during repair
Deamination: removal of an amino (-NH2) group
can change C to U
after replication, normal C-G pair becomes an A-t pair
bc there is no U in dna, it becomes a T when replicated and A is then paired w it
which types of muts do depurination and deamination result in
substitution muts
what are natural muts
mistakes in dna sequence that occur at a low freq all the time
natural processes
what are induced muts
any physical or chem agent that raises the freq of muts above the spontaneous rate
(eg x-rays, some viruses, etc)
what can excess active oxygen species in cells lead to
oxygenation of nucleotides → O2 binds to guanine, makes it able to pair with adenosine, turns that bp from G-C to A-T
substitution mutation
what do x-rays to do dna
break the dna backbone (results in deletion, translocation (swapping positions w another broken segment), or inversion)
what does UV light do to dna segments
produces thymine dimers (creates bond between 2 thymines, changes double helix structure, dna pol cant work on that region anymore :. transcription j stops, dna cant be duplicated)
t/f: all mistakes in DNA become integrated into new cells after replication
false → dna repair mechanisms fix most mistakes before they can be replicated
what is the first step in resolving mistakes in DNA? explain it
proofreading
proofreading portion of DNA pol is a 3’ to 5’ exonuclease
fixes most mistakes made by pol → reducing error rate to be even less than usual
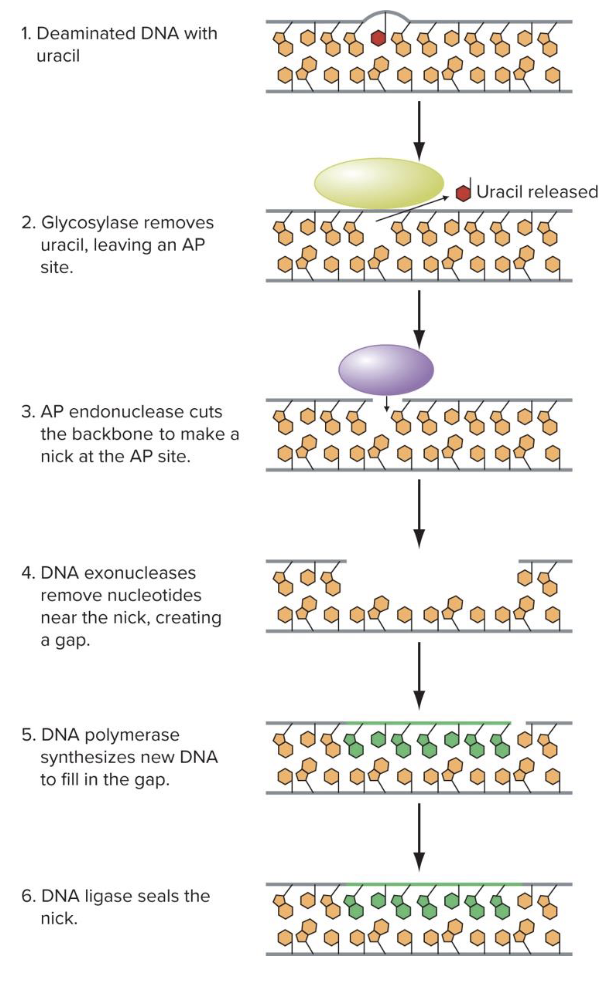
what does base excision repair do? how does it do that
removes lesions of damaged DNA
DNA glycosylases removes altered nitrogenous base and therefore nucleotides
new DNA synthesizes to fill in the gap
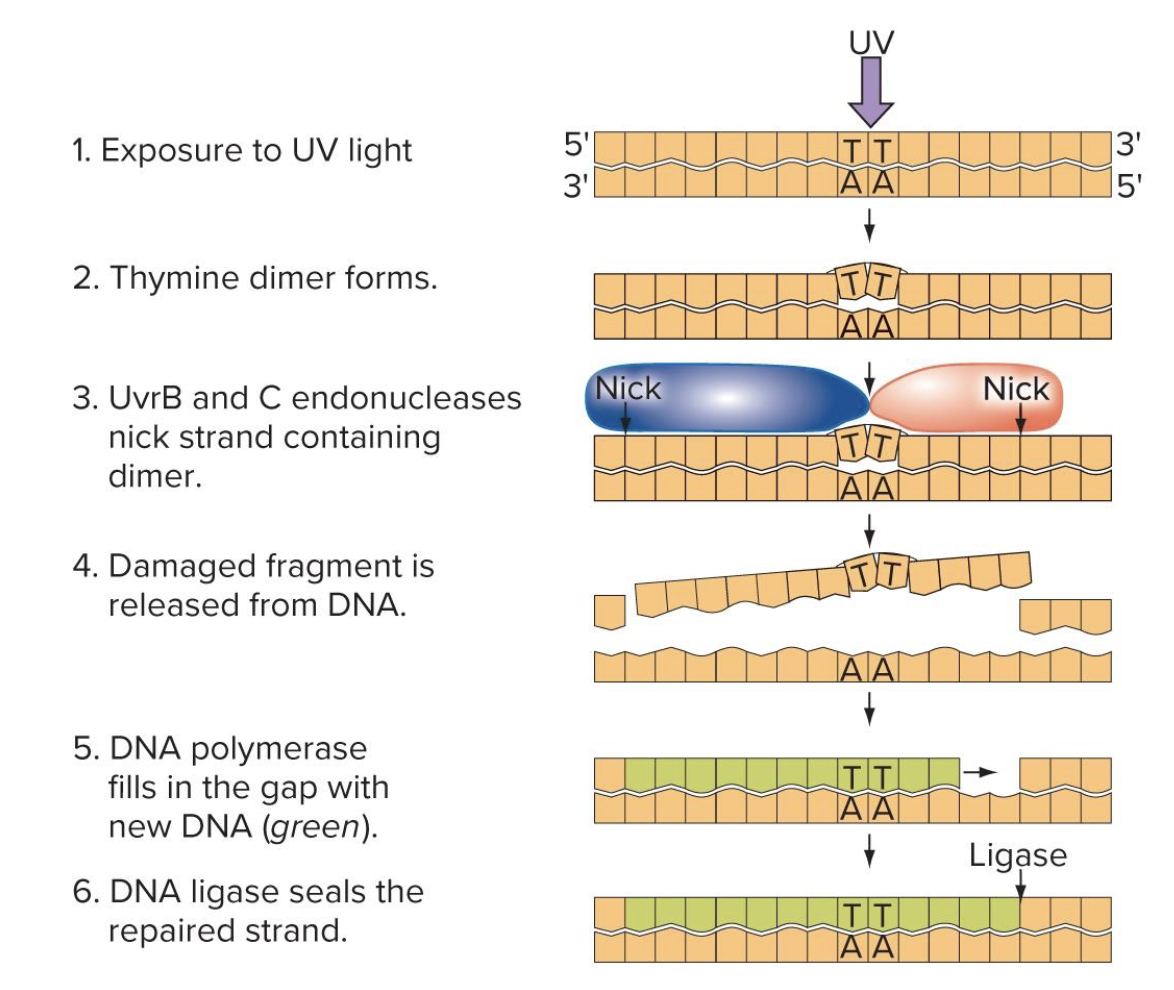
what does nucleotide excision repair do
corrects damaged nucleotides
UvrA and UvrB complexes → scan for distortions in the double helix (eg those produced by thymine dimers)
UvrB and UvrC complex → cut around the damaged DNA
dna pol fills in the gaps
what can unrepaired dsDNA breaks lead to
deletions and chrom rearrangements
what are homologous recombination and non-homologous end-joining (NHEJ)
the two ways dsDNA breaks are repaired
active during all parts of the cell cycle (not just dna rep)
two repair mechanisms
homologous recombination: similar process to crossing over during meiosis
non-homologous end-jining (NHEJ): repair of double-strand breaks without using a template
t/f: larger breaks in dna inc chances there is still a problem after the dna is repaired
true
what is the SOS system in bacteria and microhomology-mediated end-joining (MMEJ)
the last resorts to fix mistakes during replications → the error prone repair syetsms
they will introduce errors but not MASSIVE ones
SOS system in bacteria
used at replication forks that stalled bc of unrepaired dna damage
“sloppy” dna pol used instead of normal pol
adds random nucs opposite to damaged bases
Microhomology-mediated end-joining (MMEJ)
similar to NHEJ but nucs are removed at dsDNA breaks to prod short complementary regions, leading to small deletions
if mistakes aren’t caught during dna replication, how do repair enzymes know which strand is wrong
Methyl-directed mismatch repair
what is Methyl-directed mismatch repair
bacterial recognition and repair system that can fix muts AFTER dna rep
relies on “tagging” parental strands w methyl groups
euk cells also have a mismatch repair system but the “tag” is not known yet
what happens is there is too much damage to a cell at once
apoptosis → programmed cell death
too much damage to repair
eg epithelial cells dying after too much UV exposure (sun burn, peeling)
some muts derail apop so some damaged cells don’t die (common in cancer cells)
what is xeroderma pigmentosum
mut in any one of the seven genes involved in nuc excision repair
what are hereditary forms of colorectal cancer
muts in mismatch repair genes
what are hereditary forms of breast cancer
muts in BCRA1 and BCRA2 involved in dsDNA break repair by homologous recombination