Dia-MCB181 Exam 3
1/207
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
208 Terms
Binding with a small molecule results in a conformational change in one part of the protein that initiates a chain of interactions and alters its structure. These changes may conceal amino acid side chains responsible for the original DNA-binding specificity and expose a new combination of amino acid side chains that a have a different DNA-binding specificity.
How can the binding of a molecule as small as a single amino acid change the ability of a large, multi-subunit protein molecule to bind a specific DNA sequence?
A mutation in this means that the repressor will always be able to bind with the lactose operon promoter. This will lead to a cell that is unable to produce b-galactosidase with or without lactose. Lactose operon will not be inducible.
What would be the consequence a mutation in the lacI repressor gene that produces repressor protein that is able to bind to the operator but is not able to able to bind allolactose?
-helps regulate the order in which successive carbs are utilized by the bacterium.
-in the absence of glucose, cAMP levels are high. CRP-cAmp binds to a site near the promoter of the lac operon and activates transcription
-with glucose, cAMP levels are low, CRP-cAMP is not formed and cannot bind so transcription is not induced to high levels
What is the role of the CRP-cAMP complex in positive regulation of the lactose operon in E.COLI?
Lactose Operon
a protein that activates gene expression upon binding DNA found specifically in E Coli
CRP-cAMP complex
a positive regulator of the lactose operon
A mutation in lacl does:
not produces a functional repressor
the lac genes are expressed in the presence and absence of lactose
One mutant and one non-mutant lacI does:
expressed lac genes only in the presence of lactose
A mutation in lacO does:
presents repressor protein from binding so the lac genes are expressed in the presence and absence of lactose and have constitutive expression.
One mutant and one non-mutant of lacO does:
lac genes are expressed in the presence and absence of lactose and have constitutive expression
Binds to operator
What do repressor proteins to?
lacI
Produces a mRNA that encodes a protein called the lactose repressor protein
Is outside of the lac operon
lacZ
Encodes enzymes needed to split lactose into smaller sugars
-produces B-galactosidase
lacY
encodes a membrane protein needed to transport lactose into the cell
-produces lactose permease
no repressor= transcription occurs
What happens when theres lactose present?
it is induced. The operator is not bound with the repressor, the promoter recruits the RNA polymerase complex and transcription of the polycistronic mRNA occurs.
in the presence of lactose, what happens to lactose operon?
Operon
the region of DNA consiting of the promoter, the operator, and the coding sequence for the genes
found in bacteria/archae/ primarily in prokaryotes
A set of multiple bacterial genes that are regulated together and are trasncribed into a single mRNA (they share a promoter)
The part of the mRNA corresponding with each gene is translated into the protein encoded by that gene
Usually contain multiple genes that functionally reglated proteins (they all contribute to the same biochemical/cellular process) and transcribed together
these cells can translate operon correctly due to their ribososomes ability to initiate translation anywhere along an mRNA that contains a proper ribosome-binding site.
Why do operon only found in bacteria and arcahe
Cistron
coding sequence
Polycistronic RNA
a single molecule of mRNA located next to one another along bacterial DNA
true
true or false: lacz and lac y are not transcribed together
Operator/ Lac O
the binding site for the repressor and its a sequence
between lacI and lacZ
Where do lacY, promoter, lacP and an operator located?
lacP
function is to recruit the RNA polymerase complex and initiate transcription
Structural gene
they ccode for the sequence of amino acids making up the primary structure of each protein
Trytophan
when sufficient, it binds with a regulatory protein to form the functional repressor, and the transcription of the genes does not occur
when drops too low, repressor does not form, transcription is initiated.
Positive Control
the abilitiy of the pressor to bind DNA OFTEN DETERMINED BY An allosteric interaction with a small molecule
Repressor
preventing transcription of the genes into messenger RNA.
Allosteric Effect
a change in the activity so the protein can bind to the molecule with the activator with the small molecule
5’ TATAA- 3’
helps recruit proteins needed for trasncription
negative regulation
must bind to the DNA at a site near the gene in order for transcription to be prevented.
transcription takes place at a constant rate unless something turns it off
Transcriptional Regulation
the mechanisms that collectively regulate whenever transcription occus
can be positive or negative
Positive Regulation
protein must bind to the DNA at a site near the gene in oorder for a transcription to take place
Transcription is stimulated
DNA Sequence in
eukaryotes: Enhancer
Prokaryotes: Activator Sequence
Primary transcript
the initial RNA transcript of a gene is itself the mRNA immediately combines with ribosomes to undergo translation into protein
Gene Expression
the readout of genetic information flow in a cell that focuses on transcription, and translation that produce functional gene products.
Gene regulation
various ways in which cells can contol gene expression
can occur at any steps, even after the protein is made
Eukaryotes vs Prokaryote gene expression
In eukaryotes, it can be regulated at the level of chromosome packaging which involves the modification of histones, nucleosomes and DNA. They also have multiple enhancers
in prokaryotes, it can be regulated at the level of transcription and environmental levels.
All cells
must be able to adjust which genes they re transcribing/translating at any given moment to respond in real time to environmental factors
Gene Regulation in Transcriptional Level
Epigenetic Regulation (make DNA sequences more/less accessible for transcription)
Binding of regulatory proteins to non-coding sequences on DNA (eg.TFs binding to enhancers in eukaryotes)
Eukaryotes
Where do gene regulation transcriptional level regulate only?
Gene Regulation Post Transcriptional Level
How mRNA is processes
Splicing
mRNA stability
only eukaryotes
Gene Regulation Translational Level
Whether initiation complex can assemble at 5’ cap
Negatively Regulated
Transcription is repressed
The organization of prokaryotic DNA is not packaged in chromatin, mRNA is not process, and no nuclear membrane seperating the processes of transcription and translation
Why is gene expression in general simpler than eukaryotes?
Metabolizing Glucose
provides greatest net energy “pay off”
E. coli can turn lactose into glucose and galactose
If glucose isn’t available:
Lac Operon
Purpose: to avoid wasting the energy and materials for making lactose metabolism proteins if they aren’t needed, E. coli can regulate the expression of the genes encoding these protein
contains 3 genes that code for 3 proteins involved in lactose metabolism
Anatomy of Operon
DNA-operon starts at promoter-operator-structural genes(A-B-C-D) operon ends
lacz, lac, y , lac A
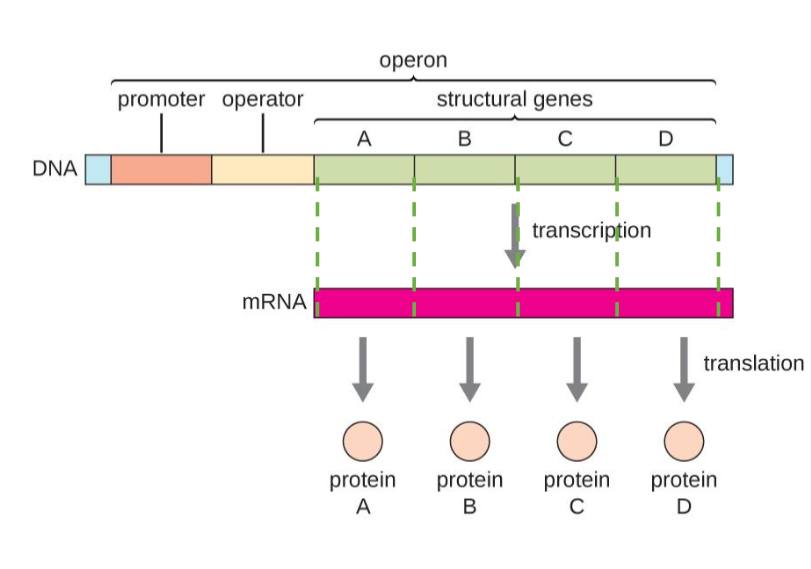
LacA
codes for another protein that lac operon contains
lacZ, lacY, lacA
What are the 3 codes that is involved in lactose metabolism?
lacP
Controls the transcription lacZ, lacY, lacA.
It eats it and break it down for energy
What do E-Coli do to lactose sugar?
E-Coli turns on genes that code for proteins
What happens if the lactose is present in the environment?
E-coli keep these lactose metabolism gene off to avoid wasting energy.
During these times, cells don’t need lactose metabolism proteins.
LacI is constitutively continually and constantly expressed
If lactose is not present?
Lactose
Indirectly unrepresses lac operon transcription
Activating proteins factors
bind to genes’ enhancers
General transcription Factor
bind to the DNA at the promoter
Specific TF Proteinns
recognizze specific DNA sequences (enhancers and promoters)
Mediator Complex
This is recruited when all TFs are bounded and then DNA loops
Connects enhancers TFs and General Tfs and the promoter
Enhancers
influence on whether, where, when, and how much a promoters it attached to and how it’s going to be active
Promoter
Acts like a switch on whether transcription would occurs or nah
The sequence of DNA where RNA polymerase binds and begins transcription of the gene
Silencers
Represses transcription (negatively regulated)
Combinational Control
The different possible combination of various TFs bound to their respective regulatory regions for a gene provide multiple layers of fine-tuning, the timing, and speed of the gene’s transcription
The regulation of gene trasncription by means of multiple TFs acting together
Mutations
In regulatory, non-coding regions of DNA affects levels of transcription
Epigenetic Regulation
Changes in how DNA is packaged in the nucleus that affect gene transcription
changes of bases
chemical modification of histones
chemical modificaiton of DNA
the way in which DNA is packed within chromosomes/ alterations in chromatin structure / chromatin remodeling
only occurs in Eukaryotes
Chromatin
the complex of DNA and proteins that forms chromosomes. this is also The DNA that exists in a cell when it is not dividing is a thread-like structure formed by wrapping DNA around histones.
Hereditary molecule wound around histones
Histones
in Eukaryotes, the proteins around which DNA is wound in each nucleosomes
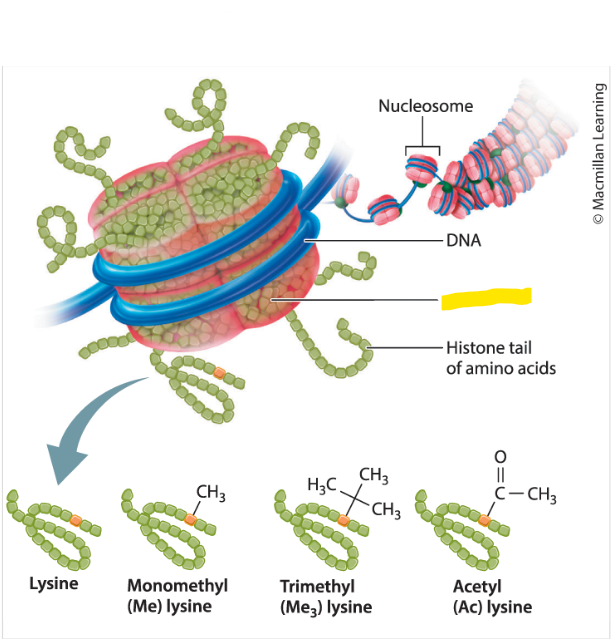
Ways that a Eukaryotic Cells can control whether and how it uses a gene in the DNA to make a functional protein
Controlling which transcriptions factors are available in the cell to bind to the genes regulatory sequences
alternatively splicing the mRNA trasncript
controlling the stability of the mRNA transcript
blocking initiation factors from binding to the 5’ cap of the mRNA transcript
modifying the protein product after translation to change its activity
B- Galactiosidase
the enzyme that breaks down lactose to galactose and glucose
Permease
Allow genes to enter the cell and facilitate transportation between the cell membrane organelles
Positive Regulation of the Lac Operon
A combination of unblocking and activating transcription
Whether and how much a gene is transcribed
depends on the exact combination of regulatory TFs and much they bound to their respective regulatory DNA sites
Alternative Splicing and RNA editing
How can one protein-coding gene code for more than one polypeptide chain
through the action of small regulatory RNAs that can either inhibit translation of degrade mRNA
Altering 5’ cap of the mRNA, so trnaslation initiation cannot occus
altering or deleting the poly(A) tail, which inhibits translation
What are three ways in which gene expression can be influence after mRNA is processed and leaves the nucleus?
Regulatory Transcription Factors
A protein that recruits the components of the transcription complex to the gene
Highly diverse and specific to one or a subset of protein coding genes
Mostly bind to enhancers
General Transcription Factors
Assemble near a short sequence in the promoted called the TATA box, the same set of protein for all protein-coding genes
RNA Processing
Primary transcript being modified several times and these modification include the 5’ cap and a string of about 250 adenosine nucleotides to the 3’ end to form the poly(A) tail
Poly A tail
helps to determine how long RNA will persist in the cytoplasm before being degraded. This also where translation is enhanced that bring the tail into contact w/ the initiation machinery on the 5’ Cap
Spliceosome
a biochemical machinery that exons are being in one during RNA splicings. The same sequences that are recognizd as exon can be recognized as intron in other RNA molecules.
RNA editing
the process in which some RNA molecules become a substrate for enzmes that modify particular bases in the RNA. Changings its sequence and what it codes for.
mRNA molecules
-they have 5’ cap, 5’ untranslated 5’ UTR, open reading frame (ORF), 3’ untranslated region, poly- A tail.
Open Reading Frame (ORF)
containing the codons that determine the amino acids/ sequences of the protein
5’ UTR
where proteins bind to transport mRNA to particular location in the cell or can affect translation initiation directly
3’ UTR
where translations are regulated
5’ and 3’ UTR
they contain regions that bind with regulatory proteins. These RNA binding proteins help control mRNA translation and degradation
Translation Initiation Complex
While some mRNA from a gene are being translated, others transcribed may not have these.
3’ UTR and Poly A Tail
initiation of translation
Post-translational Modification
Proteins are modified after translation in ways that regulate their structure and function. They help regulate activity and can alter protein folding and target molecules to particular cellular compartments.
Chaperones
Protein that help in proper folding
Different Level of Genes
Genes regulation can occur at any one or more of the steps of the gene expression: Chromatin, transcription, RNA processing, translation and post-translational modification.
Interphase and M-Phase
2 phases of cell cycles
Interphase
a process of growth and replication. Has 3 subphases. Moreover, during this, parent cell’s DNA must be faithfully replicated so that when the cell divides in two during M-phase, each daughter cell gets the exact genetic info.
G1, S1 and G2 phase
Interphase subphases
G1 Phase
-Increase volume of cytoplasm
-synthesizing new/more proteins
-producing more organelles
-building more plasma membrane to fit around all the stuff above
-Preparation of s-phase DNA synthesis
-in this checkpoint, p53 is involved
S Phase
entire DNA content in nucleus is replicated or where DNA synthesize. This is where DNA is being replicated
-this is when 46 total chromosomes gets replicated and cell divides in two, each new cell identical copies of these 2 sets chromosomes 1-23
G0
Phase
Cell that are not actively dividing
G2 Phase
there are more growth and prepares for mitosis and cytokinesis (M-phase)
Both the size and protein content increase in size
in this checkpoint, it ensures DNA replication was successful and is complete
Complementary Daughter Strands
They are not synthesized the same way on both DNA templates (parents strands)
are antiparallel to their respective parent templates
their cells has a complete copy of DNA strand this means that the chromosome stays the same in mitosis
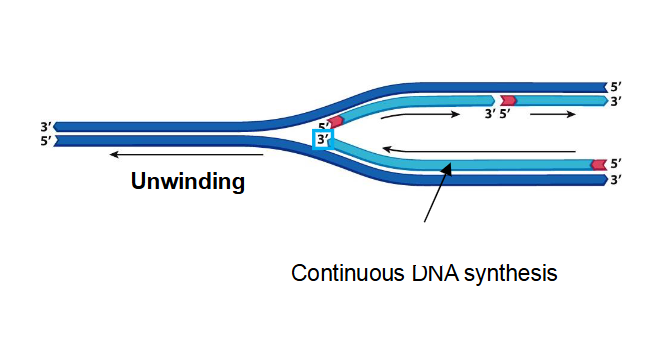
Leading Strand
The daughter strand with its 3’ end pointed towards replication fork, can be synthesized continuously
they fray from 1 primer that is required to start synthesis
Being added in the same direction as the replication fork opening