ANHB3323 epigenetics
1/14
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
15 Terms
What is epigenetics?
Study of heritable change si gene expression that occur without directly altering the underlying DNA sequence.
DNA is wrapped around histones
nucleosomes are strung together
comprises chromatin fibre
how packed chromatin in affects how it is read
epigenetics are ADDED or REMOVED mods for how genes are read (reversible)
Explain gene expression and chromatin¤
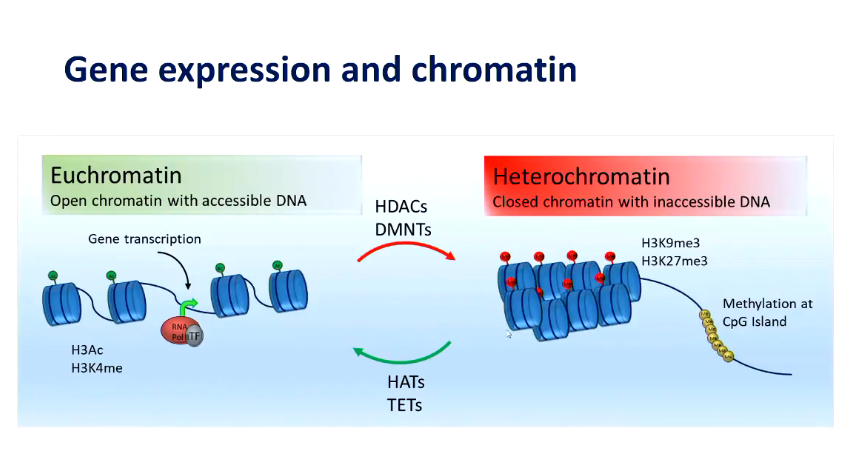
Epigenetic processes
Main two:
DNA methylation
Histon emodification
Also non-coding RNAs (ncRNAs) - anything that controls chromatin structure
Chemical modificaiton to DNA - DNA methylation
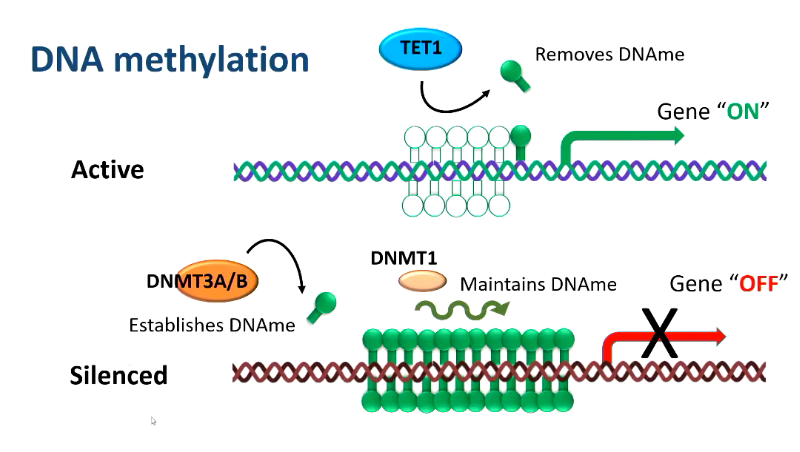
DNA methylation
heritable mark
covalent transfer of methylg group to cytosine ring
typically at CpG dinucleotine (cytosine-phosphate-guanine)
in the context of promoter region - increased methylation —> gene repression
DNA methyltransferased DNMT3A and DNMT3B establish methylation patterns in combination with DNMT3L
DNMT1: maintainence methyltransferase
these enzymes control addition or removal
Chemical modifications - histone modification
post-translational mod of histone proteins
methylation, phosphorylation, acetylation, ubiquitylation and sumoylation
impact chromatin structure
affect histone-DNA or histone-histone interacrions
recruit histone modifiers
histone modificaiton occuring can recruit more modifiers
Histone acetylation
tails produce off of histone proteins
writers of acetylation - histone acetyltransferases enzymatically add acetyl group
erasers- histone deacetylases remove acetyl group
acetylation = active gene regions
deacetylaiton = repressed
histone methylation
more stable than acetylation
occurs on lysine and arginie residues on histone tales
more complex: mono-, di-, tri-meethyl
generally associated with gene reprsioon - sometimes activation
histone demethylases - add group
histone methyltransferase - remove group
Non-coding RNA
RNA that do not have role in coding protein
long nCRNAs = >200 nucleotides and up to 100 kilobases
short ncRNAs = <200 (microRNAs most studied in cancer)
microRNA small single stranded: 18-22bp length
can drive sequence for degradation
sponging miRNA can prevent lncRNA from modifying
lncRNA can act as decoy to bind to marker instead of the marker binding to DNA
Cancer
epigenetic and genetic alterations affect phenotypic plasticity
cancer cell heterogeneity —> spectrum of cellular behaviours
^contributed heavily to resistance
epigenetic landscape altered in cancer - involcing epigenetic alterations to DNAmethylaiton and histone modification
spectrum of cell s(Eg. ‘“ïnbetween cells”’ not mesenchymal and not epithelial)
Role of epigenetics in cancer
organisation of chromatin structure differs between cell types
in cancer cells the organisation is similar to stem cells or reprogrammed cells
tumour supressor genes are repressed - inactive, mutated or silaced or deleted relative to normal tissue
oncogneic genes - overexpressed or overactive: maintain uncontrolle dporliferation, active migrationa nd invasion, promote drug resistance
Abnormal epigenetics in cancer
tumour cells have different “methylome”compared to normal cells
demethylation of promoter region sof oncogenes
OR increased in focal methylaiton also obsrved - methylation at promoter region of tumour supressor cells
Epithelial to mesenchymal transition
In normal conditions there is increased pro-epithlial miRNAs which keep mesenchynal trancription factors in check
upregulation of pro-epithelial markers —> give rise to non-motile peithelial cells tjat have good cell-cell adhesion an dpolarity
In cancer miRNA silenced by epigenetic markers —> transcription factors activated
lose cell-cell adhesion and polarity and become elongated so they can migrated
cells become more resistant to drugs
Epigenetics feedback loop
e-cadherin gives structure to membrane of epithelial cells - maintain phenotype
tramscription factors bind to e-boxes in front of epithelial genes —-
> can recruit epigenetic modifiers to promoter region of genescan remodel chromatin, add histone marks, add methylation or remove
can silence E-cadherin (CDH1 encodes for it)
loss of e-cad enabled tumour cells to metastisise
n-cadherin increased when e-cadherin decreased - CDH2 increases
miRNA can repress transcript of ZEB1
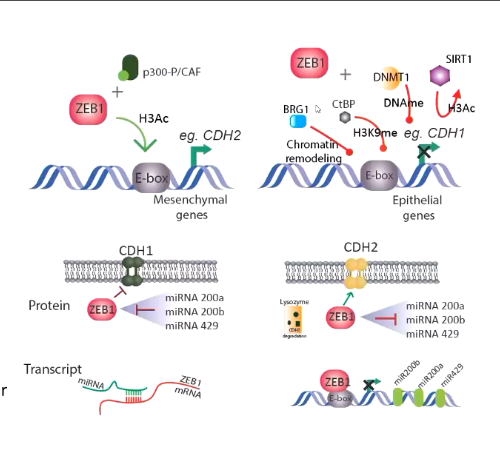
example: histone marks on oncogenic ZEB1 promoter
in non-cancer state ZEB1 is repressed by K27Me3
In cancer stem cell like state it has activaito mark of K4ME3
in hybrid state the ZEB1 promoter had both marks
bivalent mark - both on promoter
ZEB1 can switch on or off in response to environmental stimuli
epigenome editing
manipulate epigenome
3 molecular platforms developed- zinc finger proteins
simple to engineer and deliver
small - wide variety of context/use
lots of off target effects
transcription activator-like effectors
CRISPR and CRISPR-Cas