SNAREs
1/75
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
76 Terms
List the physiological systems which require membrane fusion
Discuss the experimental evidence for the role of SNAREs in membrane fusion.
Describe the key features of a SNARE molecule.
Discuss why SNARE molecules are required to drive membrane fusion.
amino acids found in high abundance in signals required to trigger active transport in the nucleus
pro, lys, arg
List some essential mechanisms carried out by membrane fusion
Synaptic vesicles fusion (communication between neurons and muscles)
Secretory granule fusion (endocrine and exocrine pancreas)
Secretion of serum proteins (albumin from hepatocytes and antibodies from plasma cells)
Mucus secretion (epithelial mucosal cells)
Intracellular transport of proteins between organelles in all of your cells
Which 2 peptide hormones are transported through secretory granule fusion ie exocytosis?
insulin and glucagon
How were secretory vesicles first identified experimentally? What was understood at this stage?
using electron microscope + stimulation of cells showed mvt of granules implying their role in the stimulated process
mechanism vas visualised but not molecularly understood
What were 3 approaches taken to identify the machinery of vesicle transport, bringing vesicle understanding from microscopic anatomy to that of the molecular machinery of vesicle fusion?
Biochemical reconstitution.
Yeast Genetics.
Cloning.
Describe biochemical reconstitution
experimental technique where a complex biological process is recreated in vitro (outside of a living cell) using a minimal set of purified components
Aim of biochemical reconstitution
identify the necessary molecules and understand the fundamental mechanisms of a process by isolating and testing the function of specific molecules by reassembling them under controlled conditions
Explain the intra-golgi transport assay
cell-free, only donor golgi with labelled mutant that cannot be modified mixed + acceptor golgi with WT + cytosol and ATP
labelled cargo will be modified by acceptor membranes
quantify transport rates, identify necessary components (like specific proteins and cytosolic factors), and test how different conditions affect the process
What was biochemical reconstitution used before looking at membrane fusion? Why was there scepticism when looking at this technique for membrane fusion?
mechanisms of ATP synthesis, DNA replication, RNA transcription, protein synthesis, the genetic code
microscopy showed membranes in very close proximity - believed that if this homogenisation was destroyed then transport would no longer happen
How was biochemical reconstitution used to understand membrane fusion?
intra-golgi transport assay showed necessary conditions to the process and what affects it eg temperature, presence of specific molecules etc
What does NSF stand for and what is its role?
N-ethylmaleimide Sensitive Factor
ATPase so hydrolises
What effect does N-ethylmaleimide have on a NSF? What is it therefore termed?
inhibits it
alkylating reagent
What does N-ethylmaleimide bind to? What does this lead to?
N-ethylmaleimide Sensitive Factor ie NSF
inhibition of NSF
What allows NSF to bind to membranes, allowing the binding of N-ethylmaleimide to inhibit a reaction?
SNAP ie soluble NSF attachment protein
How was SNAP found?
When washing membranes with salt wash - SNAP target was rinsed away and identified at the target of NSF, allowing their attachment to the membrane
What effect do SNAREs have on SNAP function? Where does the term SNARE come from?
SNAREs are proteins that allow SNAP to bind to membranes ie to allow N-ethylmaleimide to bind NSF ATPases to inhibit reactions
SNAP + REceptor
What is the role of NSF in membrane fusion?
part of a molecular machinery that builds SNAREs = membrane fusion
NSF uses ATP hydrolysis to disassemble it, resetting the SNARE proteins so they can be used again
What does SNARE stand for?
Soluble NSF Attachment Protein Receptor
How does SNAP contribute to the disassembly of SNAREs?
recruits NSF to membrane, NSF then hydrolyses SNARE complex, disassociating it to allow for their recycling
In what manner does NSF bind membranes?
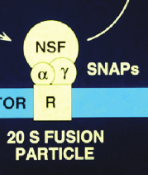
cycles on and off membranes in an ATP dependent manner at SNAP receptors, making up the 20 S fusion particle
What makes up the 20 S fusion particle ?
NSF ATPase + SNAP proteins + SNARE complex
When was the first time that it was realised that the same machinery was conserved between yeast and man?
1980 by Randy Schekman when looking at sec, NSF and SNAPs
What are alpha SNAP and sec 17 protein? What distinguishes them?
proteins binding with SNARE complexes + recruiting motor proteins NSF/Sec18
alpha SNAP and NSF in mammals // sec 17 and sec18 in yeast
What is the role of NSF/sec18?
along with alpha SNPA/sec 17, to regulate (sometimes assembly)/disassembly of cis-SNARE complexes by inducing (zippering) hydrolysis = recycling of SNARE complexes
Which gene encodes both sec 17 proteins (in yeast) and alpha SNAP (in mammals)?
sec 17 gene
Which gene encodes NSF?
sec 18 gene
What would a mutation in sec17 or sec 18 genes inhibit?
no assembly and disassembly of SNARE proteins bc non functional alpha SNAP or NSF respectively = no membrane fusion = no vesicular transport = no exchange of proteins between ER and golgi = no proteins made + accumulation of transport vesicles
What is the role of sec1 proteins?
encode Munc18 proteins that protect trans-SNARE complex from sec17/18 proteins ie alpha SNAP and NSF - preventing their disassembly and allowing it to complete assembly and mediate fusion
What does sec 1 encode in mammals?
Munc18 proteins
What are sec 1, sec 17, sec 18?
genes encoding proteins involved in membrane fusion by allowing the association or disassembly of complexes forming with SNAREs and vesicles forming and fusing
Why are pacific electric rays commonly used as models?
large neurons so good for imaging
How were clostridial neurotoxins tetanus and botulinum B used to infer the role of VAMP?
purified vesicles were taken and exposed to these toxins - showed rigidity and paralysis as VAMP disappeared → evidence VAMP is involved in membrane fusion
How were VAMP and Syntaxin discovered experimentally?
through cloning of synaptic vesicles in electric rays
What do clostridial neurotoxins tetanus and botulinum B cause?
paralysis and rigidity
What are the 3 key snare molecule identified?
VAMP, SNAP25 and syntaxin
How were the components of SNARE complexes first purified?
biochemical complementation assay - NEM treated cytosol ie NSF inactive - allowed identification, isolation and purification of non-NEM sensitive components (Jim Rothman)
What were the 4 ideas making up Rothman’s SNARE hypothesis?
1) SNAREs for each transport step within the cell.
2) SNAREs should provide specificity to vesicle transport. (not entirely true)
3) SNAREs should be sufficient to drive lipid bilayer fusion.
4) Proposed that NSF and ATP hydrolysis catalyses membrane fusion (this bit is wrong!)
Breaking down Rothman’s SNARE hypothesis, is the first part true, ie Are there SNAREs for every transport step? What types of experimental techniques were used for this research?
yes, validated! cloning trafficking material
How many different SNAREs are encoded in the human genome? What are they involved in?
38, various transport steps and fusion reactions
How was it determined that SNARE complexes are formed by zippering in a parallel coiled coil?
through crystallisation of the purified SNARE
How does a SNARE complex form?
VAMP, Syntaxin and SNAP25 come together and zipper in a parallel coiled coil
What provides energy necessary to drive membrane fusion?
the zippering of the snare complex itself
Is bringing two membranes together and fusing them energetically favourable?
no! zippering of snare complex provides the necessary energy for membrane fusion
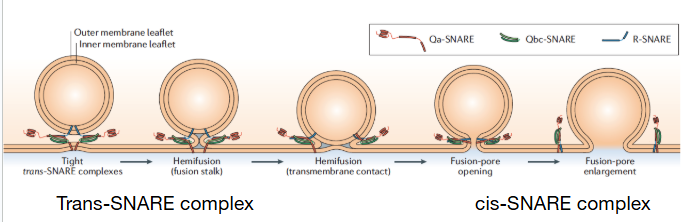
What distinguishes trans from cis SNARE complexes?
trans complex formed when SNARE proteins from different membranes pair up, fusing the membranes // snare proteins are on the same, now-fused membrane
What distinguishes R from Q SNAREs?
R snares contain an R residue, amino acid Arginine // Q ones contain a glutamine amino acid residue
Among VAMP, Syntaxin and SNAP25, which are Q and which are R SNAREs?
VAMP is R, syntaxin and SNAP25 are Q
What type of SNAREs do R-SNAREs typically act as?
arginine residue containing SNAREs usually act as v-SNAREs ie vesicle associated SNAREs
What type of SNAREs do Q-SNAREs typically act as?
glutamine residue containing SNAREs usually act as t-SNAREs ie target-membrane associated SNAREs
What is the ratio of Q to R SNAREs maintained in all complexes in vivo?
3Q:1R
How would a mutation in Q/R residues in SNAREs affect SNARE activity?
inhibits it
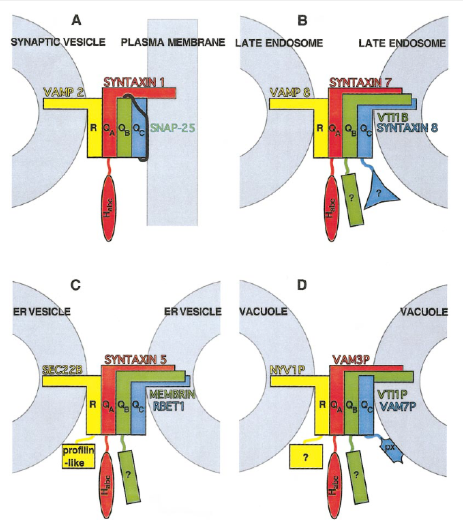
What does the following figure represent and inform on Q/R ratios maintained in all complexes in vivo?
shows various snare complexes and the snares making them up - always a 3Q to 1 R ratio maintained, no matter the different isoforms of VAMP, Syntaxin and SNAP25 involved
Breaking down Rothman’s SNARE hypothesis, is the second part true, ie Do SNAREs provide the specificity of membrane fusion?
half and half - they definitely contribute to specificity, bc even though snares show some promiscuity they mainly interact with snares from appropriate membranes HOWEVER lots of additional machinery contribute to specificty (i.e. rabs/ coat proteins and tethers) so maybe can’t say they PROVIDE the specificity
What are common features of SNARE proteins whether they have Q or R residues?
generally small 14 to 40 kDa
all have at least 1 coiled coil/SNARE motif
tm domain
c terminally anchored
What type of SNARE are Qb, Qc and Qbc SNAREs?
SNAP 25 - subfamilies of Q SNARE dependent on their location in the 4 helix bundle that SNARE complexes form
Breaking down Rothman’s SNARE hypothesis, is the third part true, ie Are SNAREs sufficient to drive lipid bilayer fusion? Which experimental technique showed this?
yes ! shown to be the minimal fusion machinery
TIRF microscopy
Breaking down Rothman’s SNARE hypothesis, is the fourth part true, ie do NSF and ATP hydrolysis catalyse membrane fusion?
WRONG
Can recombinant SNAREs fuse membranes?
yes!
What is NSF role simply in fusion?
recycles SNAREs
Via which structure do SNAREs zipper up?
via their coiled-coil domains
How are SNARE proteins generally anchored?
thanks to their C-terminal
In which fusion steps in the endocytic/biosynthetic pathways are SNAREs required?
ALL fusion steps
Which SNARE complex does NSF specifically act on?
cis-SNARE ie when fusion has occurred and SNAREs are all in the same membrane, following membrane fusion of trans SNAREs
How is NSF implicated in fusion step of membrane fusion?
isn’t! simply recycles SNAREs after fusion so essential for trafficking but not for the fusion step
Are other proteins needed alongside recombinant SNAREs to drive membrane fusion of purified liposomes?
yes! SNAREs alone can lead to membrane fusion but without calcium and Habc domains eg, much slower and would be much too slow for NTssion eg + proteins like munc 18 and complexin will provide specificity and control of fusion, NSF finally allowing resetting/recycling of SNAREs
List proteins needed in a purified liposome for successful, precise, specific and rapid membrane fusion to occur
SNAREs for membrane fusion
calcium for speed
munc18 for specificity
complexin/synaptotagmin for control
NSF for resetting and recycling
open Habc domain
How does calcium affect the fusion rate of purified membranes only in the presence of SNAREs?
increases by about 30 to 50 fold
What is the role of the Habc domain on syntaxin/Qa SNAREs in membrane fusion ?
regulates assembly of SNARE complex, acting as a lid to maintain the complex in an inactive closed conformation - inhibits unwanted fusion events. opens to allow SNARE binding
Qa SNARE
syntaxin
Qb, Qc, Qbc SNARE
SNAP25
R SNARE
VAMP
Where is the regulatory Habc domain typically found in a SNARE complex? What can this domain interact with?
at the N terminus of syntaxins/Q a SNAREs
Munc18/nsec1, ubiquitin, other SNARE domains for formation of SNARE complex
Would membrane fusion happen in the absence of calcium and presence of a closed Habc domain?
no - although snares are sufficient to fusion, calcium speeds it and Habc will typically inhibit syntaxin from binding the other snares to form the snare complex