Homeostasis, Feedback Loops, and Cellular Transport in Physiology
1/86
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
87 Terms
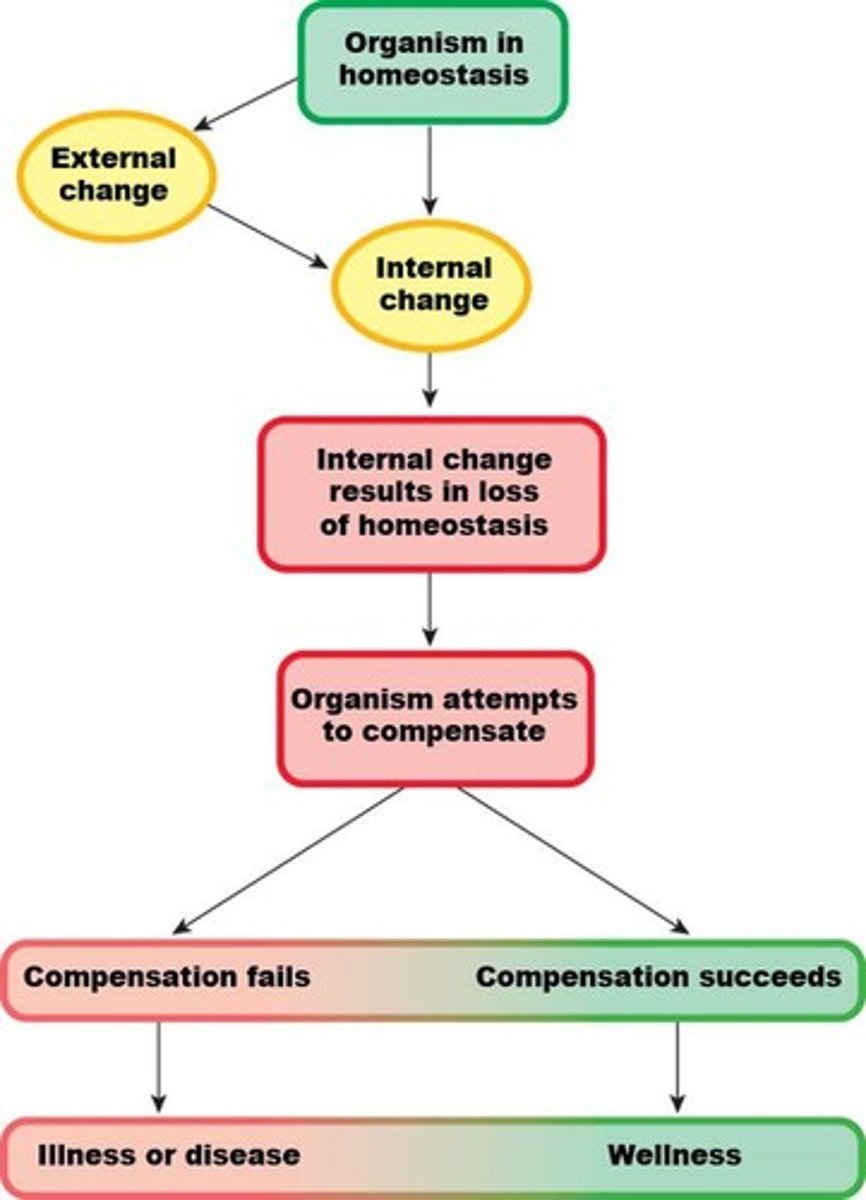
Homeostasis
The normal resting conditions of the body, including many variables such as blood glucose concentration, blood pressure, body temperature, and more.
Set-point
The normal range within which physiological variables can vary.
Receptor
Thermal receptor cells in skin that generate an electrical signal in response to stimuli.
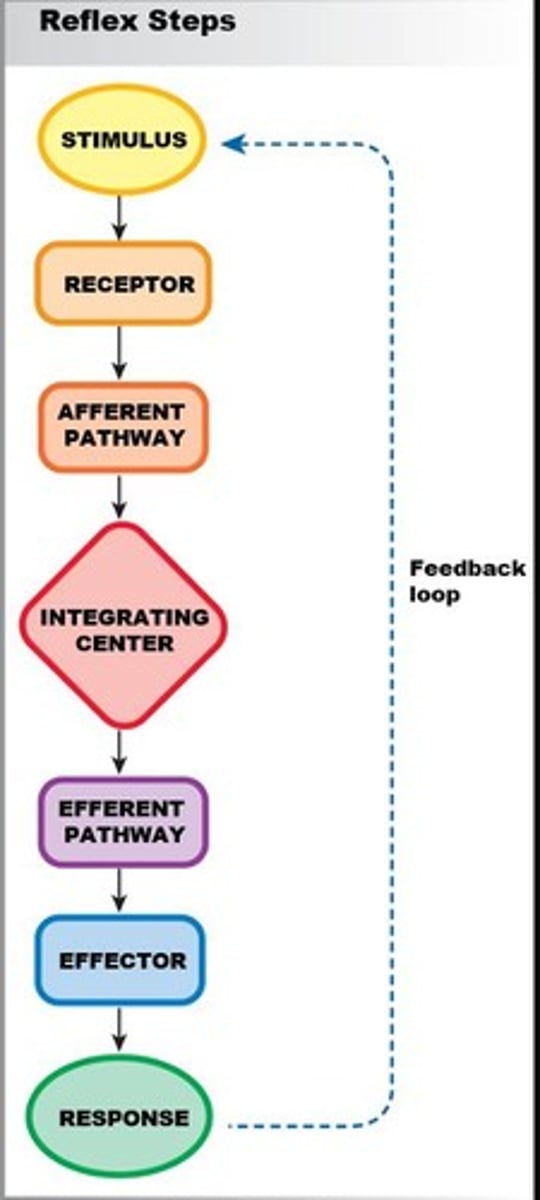
Afferent pathway
Afferent neurons (nerves) that carry electrical signals to the spinal cord.
Integrating Center
The spinal cord that processes information from receptors.
Efferent pathway
Motor neurons that carry electrical signals to muscles.
Effector
Arm muscles (flexors) that carry out the response.
Response
The action taken by effectors, such as flexors contracting.
Feedback loop
The process that results in a response to a stimulus, such as moving the hand away from a stove.
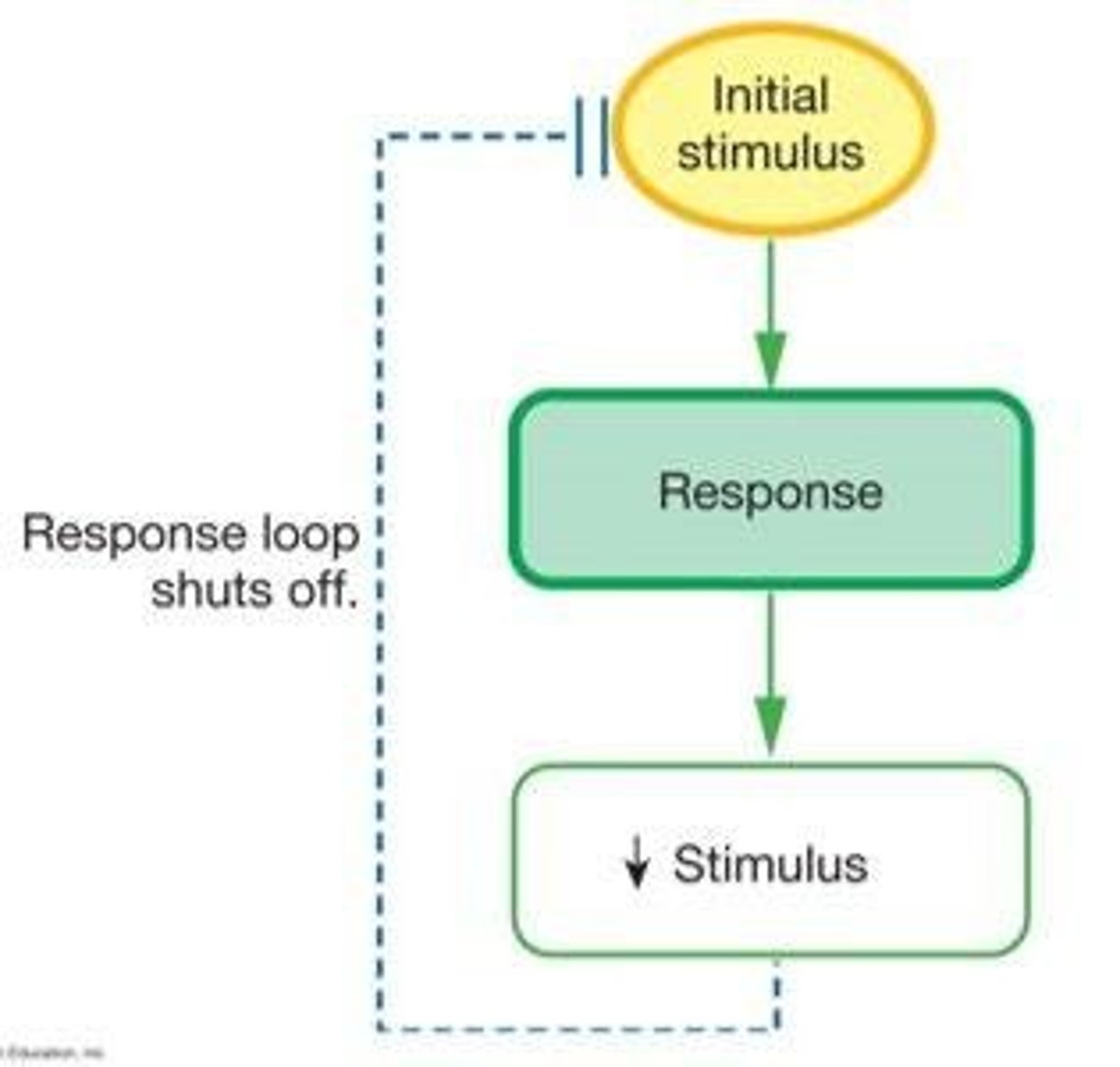
Negative Feedback
A feedback mechanism where the response is opposite to the stimulus, stabilizing a variable.
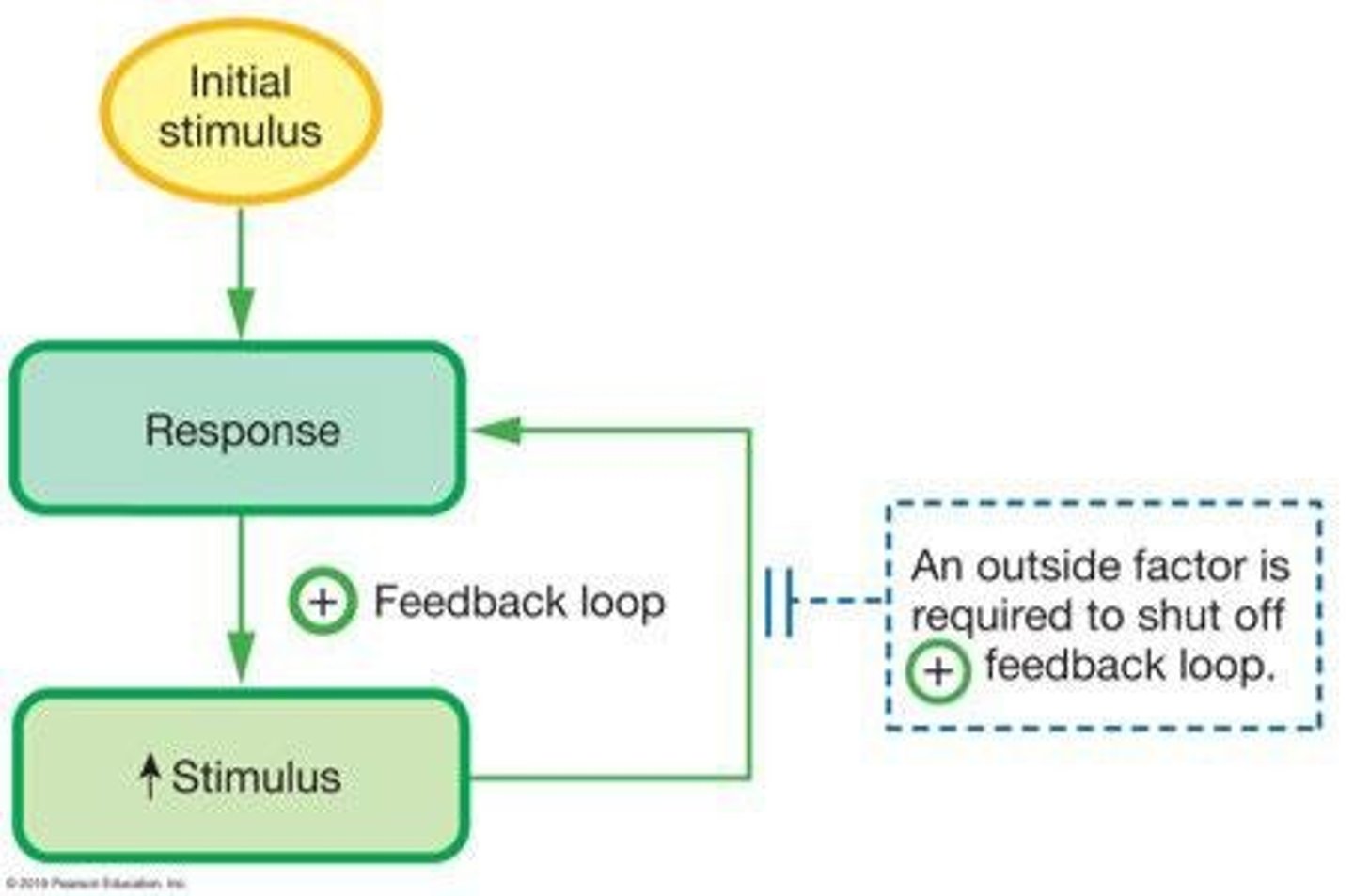
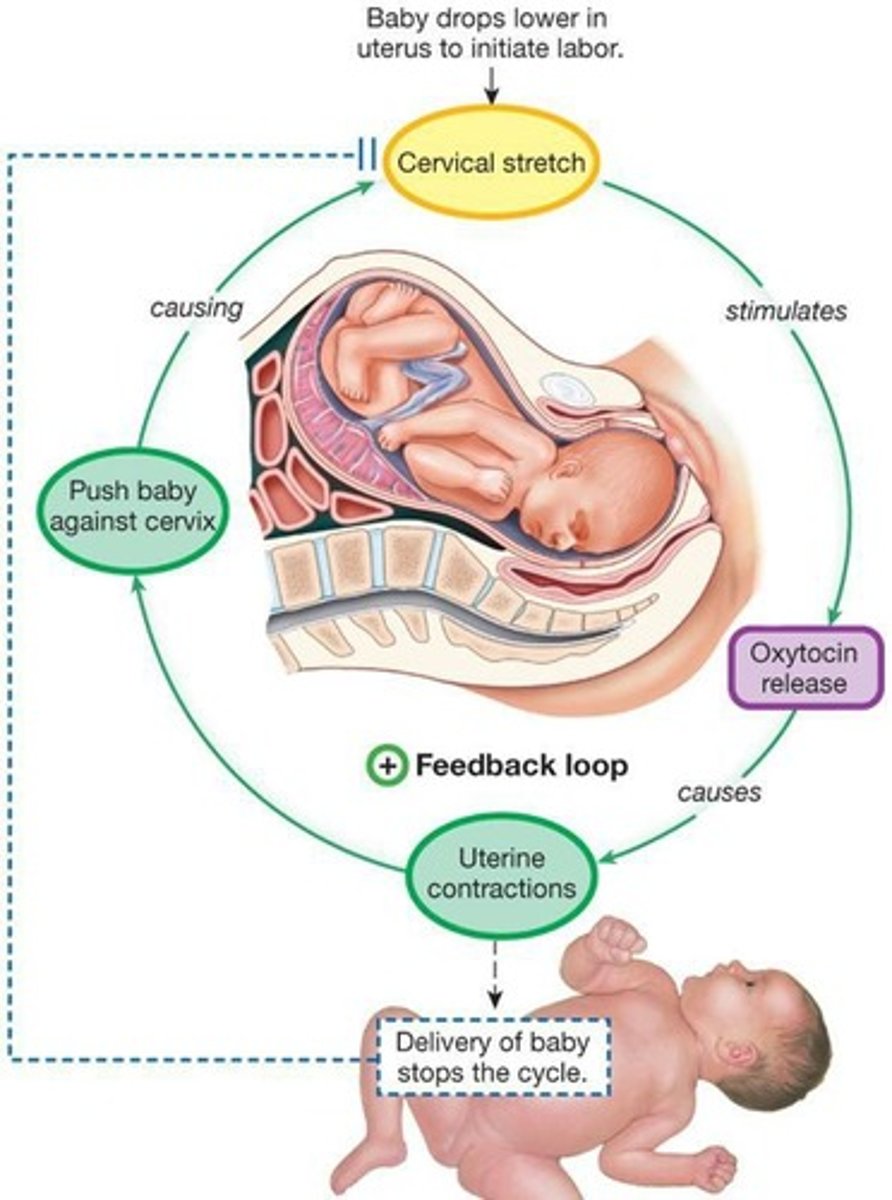
Positive Feedback
A feedback mechanism where the response is the same as the stimulus, reinforcing a change.
Feed forward control
A mechanism that allows the body to anticipate change.
Variable
A measurable factor that can change, such as blood glucose or skin temperature.
Stimulus
A change in a variable that triggers a physiological response.
Control center
The part of the body, such as the brain or spinal cord, that processes information and coordinates a response.
Afferent nerve pathway
The pathway that carries sensory information from receptors to the brain.
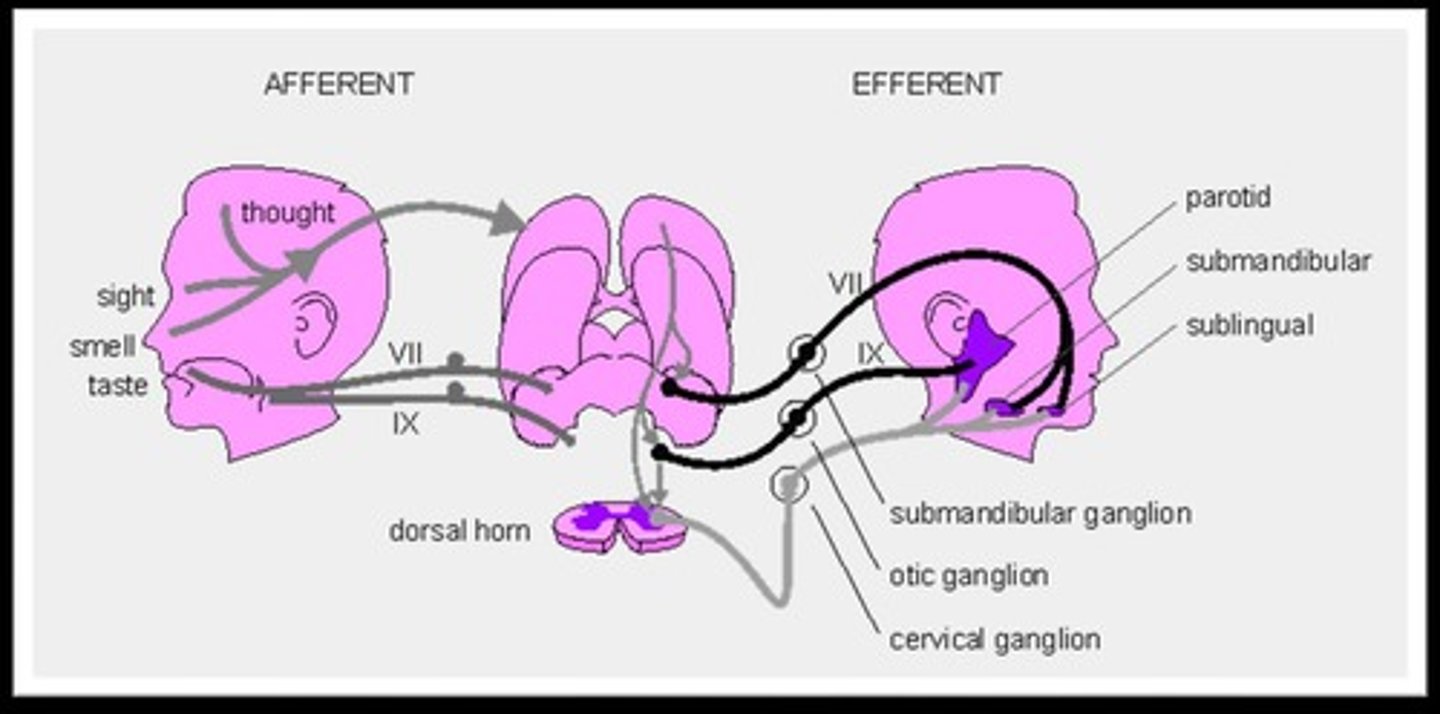
Efferent
The pathway that carries commands from the brain to effectors.
Anticipatory behaviors
Behaviors that prepare the body for a change, such as salivation in response to food cues.
Blood glucose
A variable that can increase or decrease in response to food intake.
Insulin
A hormone secreted by β-cells of the pancreas that helps reduce blood glucose levels.
Parturition
The process of giving birth, which is regulated by positive feedback mechanisms.
Saliva
A variable that increases in response to seeing, smelling, or thinking about food.
Proteins
All proteins are initially made as linear polymers of amino acids = polypeptide chain.
Amino acids
Monomers that contribute to proteins; approximately 20 different amino acids.
Peptide bond
Links two successive amino acids via the carboxyl group of one amino acid binding to the amino group of the other amino acid.
Polypeptide chain
Linear string of amino acids bound together via peptide bonds.
R-Group (side chain)
Different for each amino acid; determines specific chemical properties.
Nonpolar R-Groups
Side groups that are highly organic (hydrocarbon-based) and are hydrophobic.
Polar, uncharged R-Groups
Contain hydroxyl, sulfhydryl, or carboxamide; capable of hydrogen bonding.
Polar, charged R-Groups
Contain carboxyl, amino groups, or imidazole ring; hydrophilic and capable of ionic interactions.
Overall protein structure
Includes four levels: 1o (linear sequence), 2o (specialized regions), 3o (overall folding pattern), and 4o (multiple polypeptide chains assembled together).
Monomeric proteins
Proteins composed of a single polypeptide chain (e.g., myoglobin).
Multimeric proteins
Proteins composed of multiple polypeptide chains.
Homomeric proteins
Multimeric proteins where all subunits are the same polypeptide (e.g., LDH-1 & LDH-5).
Amino acid basic structure
General features include a central α-carbon, amino group, carboxyl group, and R-group.
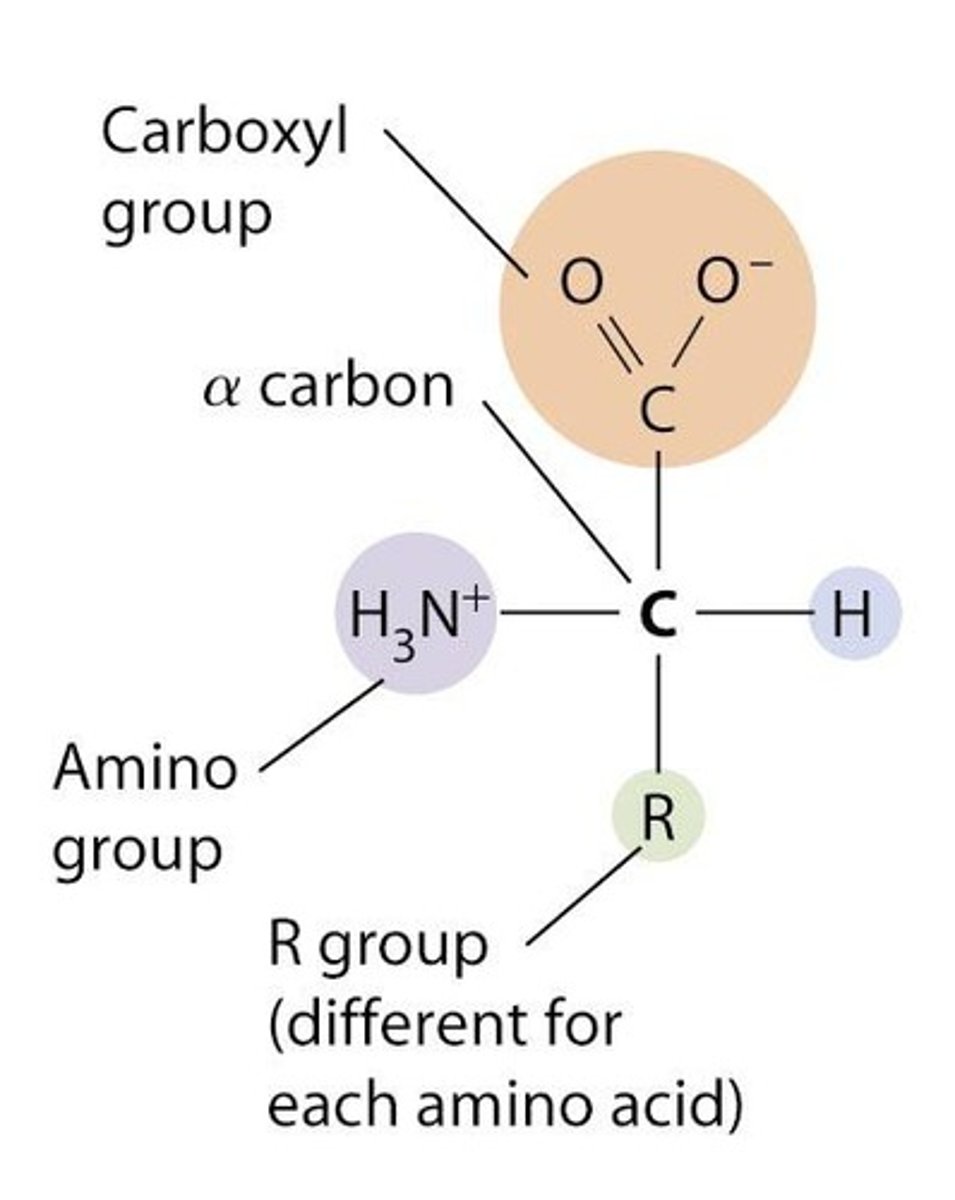
Amino acids in proteins
Most amino acids in proteins are L-amino acids.
Role of membrane permeability
Determines how substances can pass through the cell membrane.
Passive transport processes
Transport processes that do not require energy.
Active transport processes
Transport processes that require energy.
Learning Objectives
Understand the basics of protein structure, amino acids, peptide bonds, protein folding, and membrane transport.
Chapter references
Chapter 2 (pages 35-39) and Chapter 4.
Hydrophobicity
Property determined by the R-groups of amino acids.
Heteromeric
Different polypeptide chains assemble together (e.g., Myosin, hemoglobin, LDH-2, LDH-3, LDH-4)
Dimer, trimer, tetramer
Refers to the number of subunits in a protein.
Monomeric Protein
Example: Myoglobin, which consists of 1 subunit (1 single string of amino acids = polypeptide).
Heteromeric Protein
Example: Hemoglobin, which has 2 alpha (α) subunits and 2 beta (β) subunits, making it a heterotetramer.
Importance of Membrane Transport
Nutrients move into cells, wastes move out of cells, ionic balance, electrical balance (resting membrane potential), and regulatory molecules (certain hormones).
Simple Diffusion
Movement from a region of high to a region of low concentration, driven by the concentration gradient.
Facilitated Diffusion
Involves protein-mediated transport of solutes across membranes.
Active Transport
Transport process that requires energy, driven by ATP.
Direct Active Transport
Primary active transport that directly uses ATP as the energy source.
Indirect Active Transport
Secondary active transport that uses the energy from the electrochemical gradient created by primary active transport.
Osmosis
Simple diffusion of water across a semipermeable membrane, from low solute concentration to high solute concentration.
Concentration Gradient
A difference in the concentration of a substance across a space.
Mass Percentage
Mass specific particles/total mass (all particles).
Facilitated Diffusion Model
Example of simple glucose transport involving integral membrane proteins.
Carrier Proteins
Proteins that bind the solute on one side of the membrane, undergo a conformational change, and release the solute on the other side.
Channel Proteins
Proteins that form channels through which ions or molecules can pass through the membrane.
Membrane Permeability
Refers to the ability of a membrane to allow substances to pass through it.
Chemical Gradient
A difference in the concentration of a substance across a distance.
Hydrophobic Cell Membrane
Cell membrane that is impermeable to polar and charged molecules.
Equilibrium
State in which the concentrations of a substance are equal across a space.
Solute Concentration
The amount of solute present in a given volume of solution.
Integral Membrane Protein
A protein that spans the entire membrane and facilitates the transport of substances.
Uniport Transporter (Uniporter)
Transports only a single solute.
Coupled Transport
Transports 2 or more solutes.
Symport (Cotransport)
Solutes transported in the same direction.
Antiport (Exchangers)
Solutes transported in opposite directions.
Driving Force for Transport
The concentration gradient that determines the direction of movement for coupled transport.
SGLT
Sodium-dependent glucose cotransporters that are symporters.
GLUTs
Glucose transporters that are uniporters.
Na-H Exchanger
A type of antiporter that exchanges sodium ions for hydrogen ions.
Na-Ca2+ Exchanger
A type of antiporter that exchanges sodium ions for calcium ions.
Ion Channels
Channels with high specificity for particular ions, most are gated.
Ligand-gated Channels
Ion channels that open in response to a specific ligand.
Voltage-gated Channels
Ion channels that open in response to changes in membrane potential.
Mechanosensitive Channels
Ion channels that open in response to mechanical stress.
Porins
Large pores with low specificity that allow large molecules to pass.
Mitochondrial Porin (VDAC)
Allows NAD & NADH to cross the outer mitochondrial membrane.
LamB Porin
A porin in bacteria that allows maltose to enter the cell.
Aquaporins
Pores for the rapid conduction of water, found in kidney tubules.
Direct (1o) Active Transport
Transport directly linked to ATP breakdown.
Calcium Pump
Utilizes energy from ATP to pump Ca2+ across a membrane against a concentration gradient.
Na+/K+ - ATPase Pump
A direct active transport mechanism that pumps sodium out and potassium in.
Indirect (2o) Active Transport
Transport indirectly linked to an ATPase pump, using a concentration gradient.
Coupled Transporter Protein
Uses a solute gradient established by an ATPase pump to move another substance against its gradient.
Proton Gradient
A concentration gradient of protons (H+) that can drive the transport of other solutes.