BIS2A SS1 Week 5
1/90
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
91 Terms
cancer cells and telomerase
cancer cells have really high telomerase activity, so they never senesce/die and lose “important” DNA
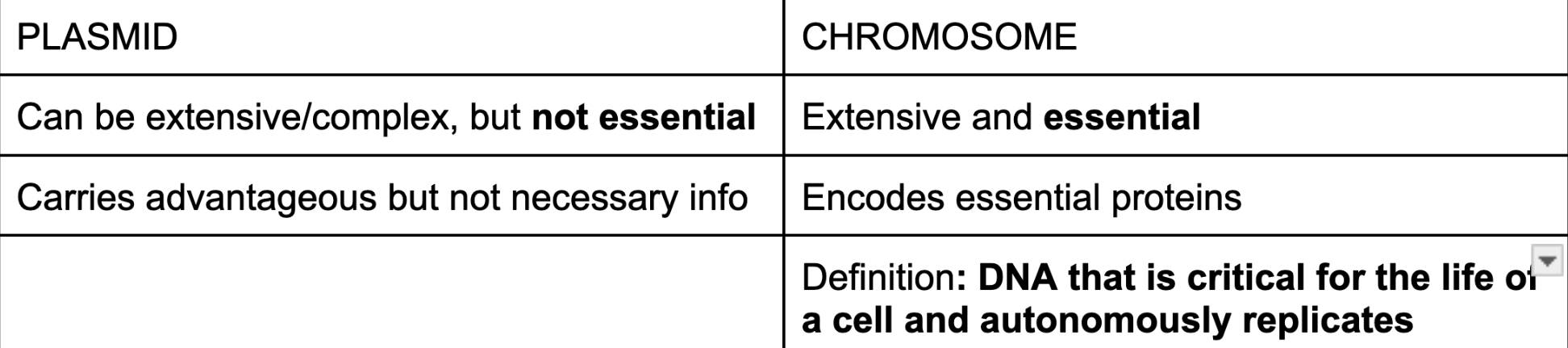
plasmid vs chromosome
what are some examples of essential genes
DNA replication/repair, membrane creation, transcription, translation, cell cycle regulating genes
3 ways that a cell can repair a mistake
repair the mistake by proofreading by pol III
DNA pol can leave the mistake behind and other enzymes can repair it
SOS polymerase in bacteria (last resort)
proofreading
when pol III detects a mistake, it can go back, cleave the bond it made, and replace the nucleotide with the correct nucleotide (exonuclease activity)
occurs in certain DNA polymerases and NO RNA polymerases
why doesn’t RNA polymerase proofread?
RNA is made and degraded, so mistakes only result in a few bad proteins
DNA mutations are permanent and can be passed down (need proofreading)
why is flu/COVID mutagenic?
they are RNA viruses, so the RNA polymerases are highly inaccurate
results in mutations in antigens to avoid immune system detections
mismatch repair (single strand repair)
replacing one wrongly inserted nucleotide with the correct one
done by DNA pol I; more local repair
excision repair (single strand repair)
removes a whole region of DNA containing mistakes and codes it again
common with frameshift mutations
done by DNA pol I
DNA methylation
used to recognize which strand is the daughter strand and which is the parent strand
can recognize which strand contains the mistake and repair it based on the methylation state
only works on repair enzymes before methylation enzyme
is the parent or daughter strand unmethylated before methylation enzyme?
daughter stand is unmethylated BEFORE the methylation enzyme comes
therefore, the mistake is in the unmethylated strand
what two processes occur if both strands are methylated and there is damage?
photoreactivation or recombination repair
photoreactivation
photoreactivation genes use the energy of the light and can leave damage cause by UV rays by cleaving C-C or T-T dimers that arise
can also repair nicks or breaks in DNA
recombination repair
using the homolog chromosome as a template for repair
eukaryotes have the advantage here as we are diploid; prokaryotes can only do this during replication
SOS polymerase
the last line of defense for bacterial cells for death, equivalent to a cell cycle stop in eukaryotes (ex. blocked replication fork)
stuffs a bunch of random nucleotides (results in a lot of mutations bc no proofreadings)
double strand breaks
when chromosomes break, need to repair the open ends
non-homologous end joining (NHEJ)
putting together breaks regardless of the chromosome (joining together the wrong chromosomes)
microhomology-mediated end joining
more homology than NHEJ but still on the wrong chromosome
homologous recombination (HR)
ends of DNA are attacked by a single strand; using homolog chromosome to repair double strand breaks
process of HR with linear into linear DNA
DNA with free ends invade (cell doesn’t like), 3’ end of that DNA is chewed up by recombination enzymes, DNA is replaced (reciprocal recombination)
2 recombination events
RecA
binds all over the DNA with free ends to align the DNA for the enzyme to complete replacement
recombination frequency
the closer genes are linked, the lower their recombination frequency is (more likely to be inherited together)
recombination is random at 50%
HR of linear into circular DNA
2 recombination events
HR of circular into circular DNA
1 recombination event
why is recombination important?
gene mapping, eukaryotic meiosis
what are the 3 ways that a bacteria can move around/swap DNA?
transduction, transformation, conjugation
transduction
phages can move DNA from one bacterial cell to another
transformation
cell can take up DNA from the outside environment (like plasmids)
conjugation
F+ (sex pilus) can replicate and transfer a plasmid to an F- bacteria (makes it F+)
central dogma
DNA —> RNA (transcription) — nucleus
RNA —> protein (translation) — cytoplasm
gene
information containing DNA that can code for proteins
transcription
DNA (gene) —> mRNA
only use a small segment of one strand of DNA (template strand)
template strand
the strand the RNA polymerase reads to make mRNA
coding/nontemplate strand
the strand complementary to the template strand
replacing the T’s with U’s gives you the mRNA
RNA polymerase
the only polymerase involved in transcription (doesn’t require primers, helicases, etc.)
in bacteria, interacts with sigma and backbone
in euks interact with transcription factors
in which way is the template strand read? the mRNA transcribed?
template strand read from 3 to 5
mRNA transcribed 5 to 3
promoter sequence
the site where the RNA polymerase binds
determines which strand of DNA is read
promoter consensus sequence
a sequence that is common throughout all bacteria, where sigma binds
sigma (bacterial intiation)
transcription factor that helps RNA polymerase bind to the promoter consensus sequence (-10, -35 promoter)
TATA box (eukaryotic/archaea promotor)
TATA binding protein binds to the TATA box, recruits enzymes to help RNA pol bind
what bases are the promoters rich in?
A and T
transcription start/initiation site
where transcription starts
overlapping/adjacent to the promoter
transcription stop/termination site
where transcription ends
what are the two types of transcription termination in bacteria?
factor independent and factor dependent termination
factor independent termination
hairpin and U sequence cause the RNA polymerase to hop off
size of hairpin + length of U sequence determine terminator effectiveness
factor dependent termination
rho protein binds to mRNA and knocks the RNA polymerase off
operons (bacteria)
polycistronic stretches of DNA, controlled by one promoter and transcription stop site
how to regulate expression of genes
adding weak terminators (hairpins) between genes allows downstream genes to be expressed less
all types of passive control of bacterial genes
promoter strength/location, termination strength/location, sigma factor
immature RNA
in euks — mRNA that is in nucleus and undergoes post-transcriptional changes to be ready for translation
what are the 3 post-transcription modification
add 5’ G cap and 3’ poly-A tail to stabilize RNA
splicing — remove introns and join exons (can make multiple proteins from same stretch of DNA)
translation
mRNA —> protein
initiation
finding the start codon after recognizing the G cap
translocation
ribosome translocates to next codon; costs 2 GTP
elongation
creating the amino acid sequence in the APE sites
termination
reading the stop codon
brings in the release factor in A site to release polypeptide chain
start codon
AUG; codes for methionine
when looking at a sequence of bases, always look for the start codon
stop/nonsense codons
stop codons don’t code for amino acids
what is needed for translation?
mRNA, amino acids, ribosome, tRNA
tRNA structure
one end has anticodons that are complementary to codons (determines if correct AA being added)
other end carries the AA
aminoacyl-tRNA synthetase
enzyme that consumes ATP to attach the amino acid to the tRNA to make it into charged tRNA
ribosome
makes proteins; composed of a small and large subunit that sandwich mRNA
made of rRNA and proteins
A site (aminoacyl site)
entry site for the charged tRNA
peptidyl site (P site)
forms the peptide bond between the amino acids
exit site (E site)
where the uncharged tRNA exits once the peptide bond is formed
ribosome binding site
the site overlapping/next to translation start on the protein-coding sequence where the ribosome binds
how much does it cost to make one peptide bond
costs 1 ATP (aminoacyl-tRNA synthetase) and 2 GTP
gene expression
the expression of genes can be regulated by a variety of factors
regulatory region
a site before the promoter that can also affect gene expression
alleles
different variants of the same gene
what prevents RNA polymerase from binding to the promoter?
1) promoter sequence
2) prevent binding with other factors
strong promoter
a promoter where RNA polymerase can bind without the help of transcription factors
weak promoter
a promoter where RNA polymerase needs help of TFs to bind
sigma protein transcription control (bacteria)
there are multiple sigma factors that control specific genes and bind RNA pol at different strengths
what promoter RNA pol binds to depends on the sigma factor and its strength
transcription factors
proteins that make the promoter site available to RNA polymerase
activators
turns on gene expression for weak promoters (positive gene regulations)
repressors
turns off gene expression for strong promoters (negative gene expression)
what are the layers of gene regulation
1) promoter strength (weak or strong)
2) transcription factors (activator or repressor)
3) regulating transcription factors
regulation of transcription factors
allosterically regulated by effector molecules
inhibitor (effector)
binds to activator to deactivate it (decreases transcription)
co-repressor (effector)
binds to repressor to activate it (decreases transcription)
inducers (effector)
binds to activator to activate it OR binds to repressor to deactivate it
increases transcription
major difference between prokaryotic and eukaryotic genomes
eukaryotic organisms often have a lot more protein targeting/secretion and cell structure genes (organelles/endomembrane system)
in humans, which types of cells do we need to be careful in replicating?
germ line cells, as they are passed down to offspring
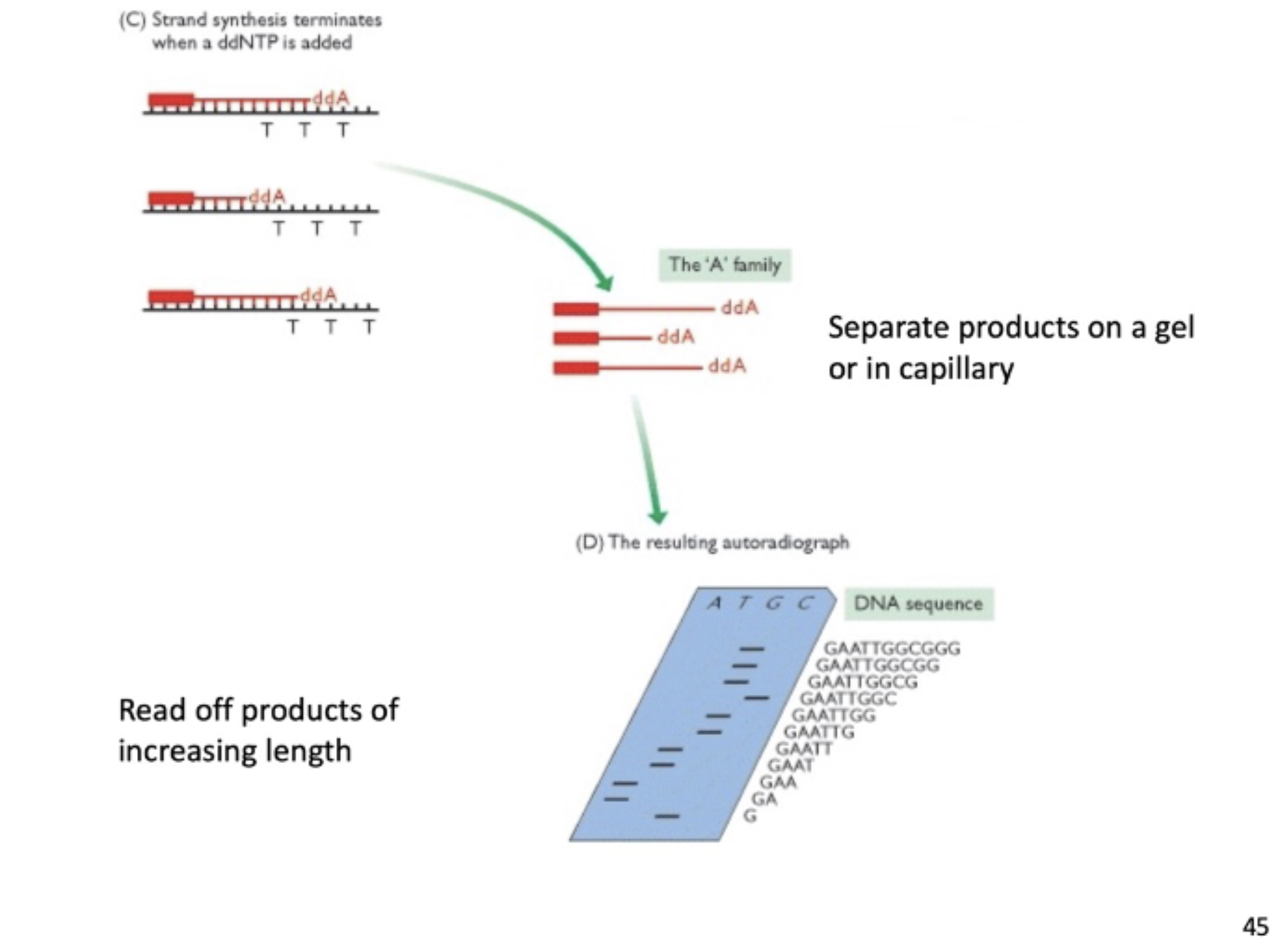
sanger sequencing
enzymatic sequencing that uses dideoxynucleoside triphosphates (ddNTP) and relies on RNA polymerase’s inaccuracy
when ddNTP is incorporated, replication stops (doesn’t have a 3’OH)
how is the DNA sequence from sanger sequencing read?
using gel electrophoresis, read off products of increasing length by nucleotide to read the sequence of the longest strand of DNA
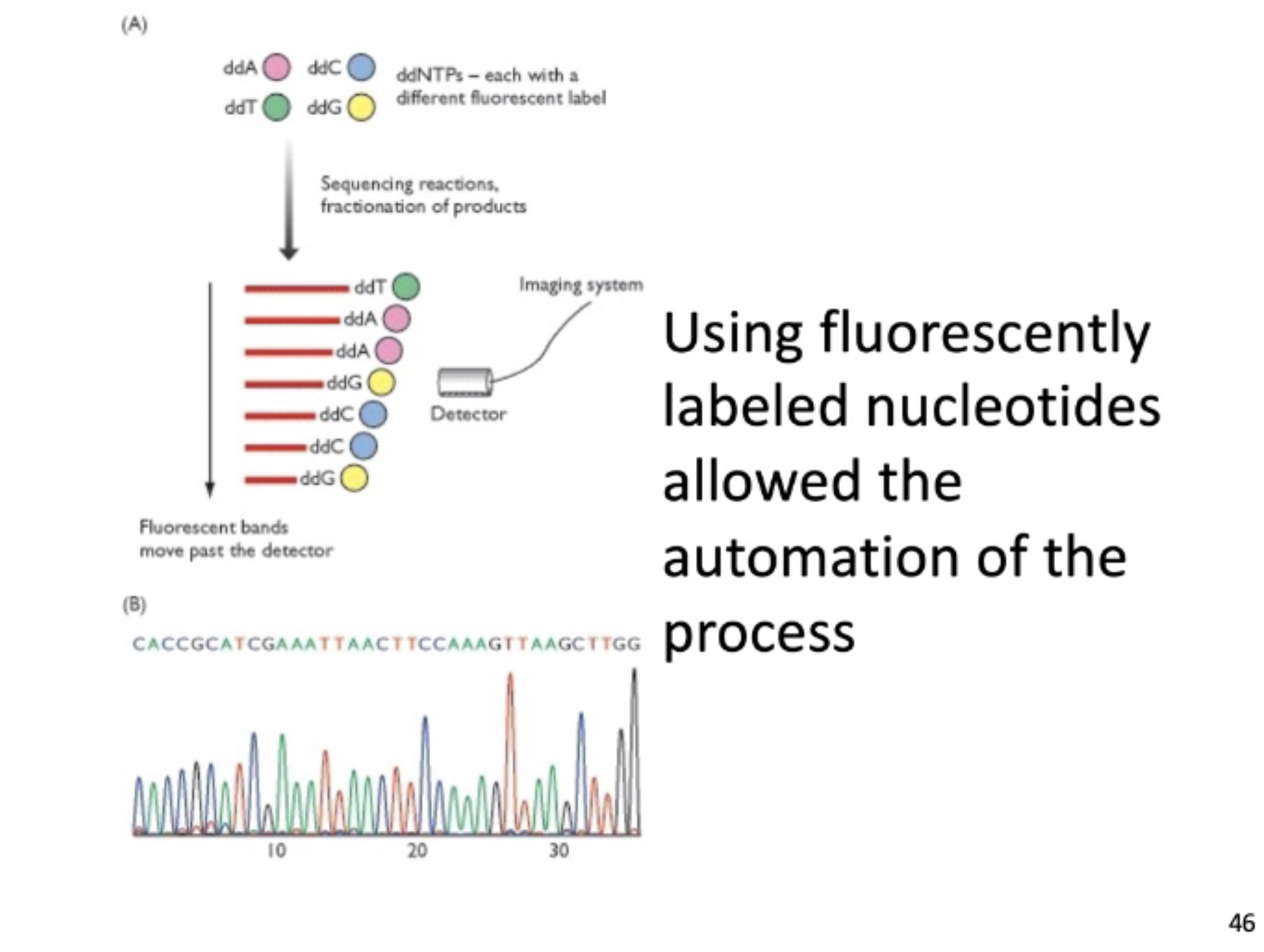
fluorescently labeled nucleotides
the more common technique, where readings of florescent nucleotides can map out the sequence
major problem with genome sequencing
works well in bacteria with only one allele
problem is repeats in sequences makes it hard to order genes and completely assemble them
BLAST
has a +1, +2, +3 reading frame, which allows you to map out the codons in DNA from 5’ to 3’
once DNA is mapped out, can compare DNA to see which protein/enzyme it is
can also do the complement strand (-1, -2, -3) because can’t determine template or coding strand
cDNA
using all message and reverse transcriptase to get all coding regions of DNA
what is the difference between a mutation in a haploid organism and a polyploid organism?
mutations in haploid organisms are much more visible in comparison to polyploid organisms
do not have chromosomes to make up for any lack of critical gene expression