enzyme regulation
1/19
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
20 Terms
list different methods of regulating e
protein expression, allosteric control, proteolytic activation/degradation, protein-protein interactions, covalent modification
summary of p reg methods to control degree of p activity
p expr - long term reg, changing transc/l of genes
allosteric control - mods bind and cause conf change that changes shape that inc or dec activity
proteo - cleaves proteins to either destroy or activate it
p-p interaxns - dis/association of reg p changes activity
covalent mods - change chemical struct of proteins by using covalently bonded groups
allosteric modulation
molec that induces change in the e conf that can change s binding, active site residue position
can be inhibitory or stimulatory, can have more than 1 mod bind
allosteric control vs inhibitors
inhibitors just turn off the e, dec [pdct] bc some e not active, not all e bind i
modulators - dec or inc activity, change catalysis to slow or quicken, does so through diff mechs, dec [pdct] by slowing e
ex of allosteric e control
asp transcarbomolyase (ATCase) - used in early step of synth UTO and CTP
CTP binds ATCase when in high concentration and turns down activity (high [CTP] = inc theta = dec ATCase activity)
s: asp and carbamoyl phosphate
allosteric e kinetics
don’t follow mm, have sigmoidal curve kinetics for V0 vs [s]
have low and high activity state
don’t have km but have k0.5, which is the [s] where V0 = 1/2Vmax (basically same as km)
± allosteric modulators
low and high activity states of allosterically controlled e
low - in low [s], t state
high - in high [s], r state
![<p>low - in low [s], t state<br>high - in high [s], r state</p>](https://knowt-user-attachments.s3.amazonaws.com/0d1aa87c-0f00-46fe-90d2-c6e7ec4451d7.png)
± am
+ / activators - s stimulates binding, dec k0.5 so takes less [s] to reach 1/2Vmax to “inc rxn vel at a fixed [substr] relative to no +am”, other kinds can inc Vmax w/o causing change in k0.5
- / deactivators - some kinds can dec Vmax w/o causing change in k0.5, some can inc k0.5 (inc amt of substr needed to reach ½Vmax) w/o changing Vmax, makes less [pdct] at similar [s], “dec rxn vel at fixed [substr[ relative to no -am”
curves more sigmoidal (+ - like r curve, - - more flat)
![<p>+ / activators - s stimulates binding, dec k0.5 so takes less [s] to reach 1/2Vmax to “inc rxn vel at a fixed [substr] relative to no +am”, other kinds can inc Vmax w/o causing change in k0.5<br>- / deactivators - some kinds can dec Vmax w/o causing change in k0.5, some can inc k0.5 (inc amt of substr needed to reach ½Vmax) w/o changing Vmax, makes less [pdct] at similar [s], “dec rxn vel at fixed [substr[ relative to no -am”<br>curves more sigmoidal (+ - like r curve, - - more flat)</p>](https://knowt-user-attachments.s3.amazonaws.com/07f079e5-8b37-4e86-b86c-2d11d4f0e600.png)
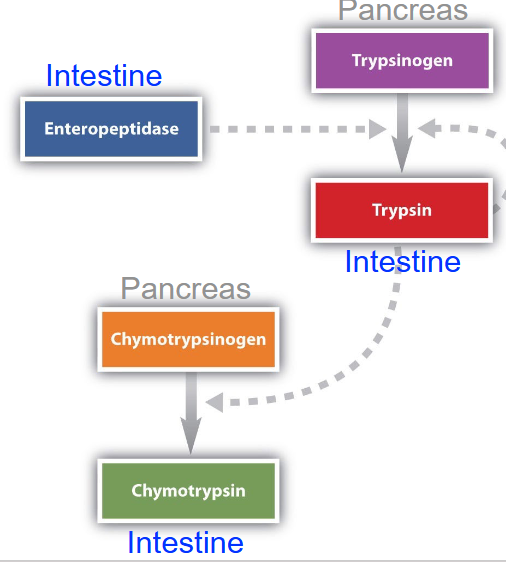
ex of proteolytic cleavage
chymotrypsin and trypsin e are made in the pancreas and secreted into the small intestine to prevent self digestion
they’re made as a long inactive recursor (zymogen) and are cleaved to become active in the small intestine (low pH)
trypsin & chymotrypsin proteolytic cleavage mech
made as trypsinogen and chymotrypsinogen in pancreas, then moved to the small intestine where enteropeptidase cleaves trypsinogen (btwn residues 6 and 7) into typsin
trypsin then cleaves chymotypsinogen into pi chymotrypsin (btwn residues 15 and 16), which leaves itself into alpha-chymotrypsin (btwn residues 13 adn 16, 146 and 149) to become fully active
pancreatitis
caused by issues with trypsinogen and chymotrypsinogen moving to small intestine bc food taken in can’t be degraded into nutrients
essentially eating but starving to death
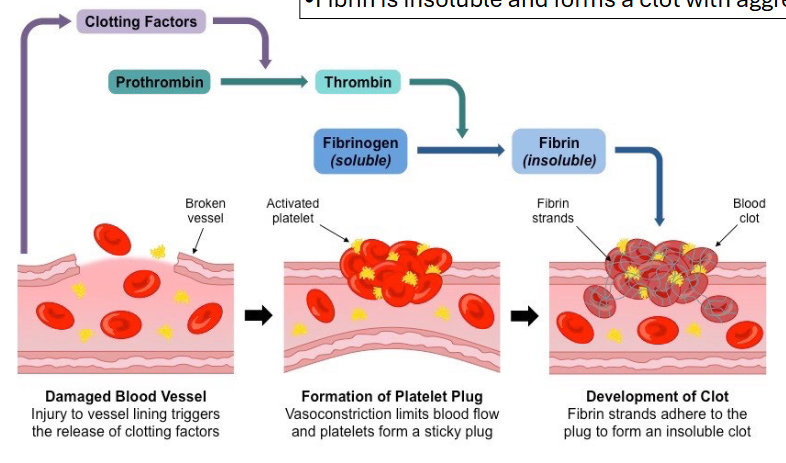
blood clotting as proteolytic cleavage reg
thrombin (ser protease) made as prothrombin (zymogen)
prothrombin hangs out in the blood and when it gets cleaved, it becomes active thrombin, which cleaves fibrinogen into fibrin
fibrin is insoluble and forms clot with aggregated red blood cells and platelets
proteolytic cleavage prevents active thrombin from being in blood all time and clotting blood all the time while allowing it to be active whenever needed
how does proteolytic cleavage help reg
allows for fast response - e are made inactive and can be turned on/off quickly when certain thing present
ex. blood clotting - fast response to injury w/o blood clotting all time bc injury to a blood vessel releases clotting factors that cleave prothrombin
ex. chymotrypsin - prevents self digestion of pancreas but allows e to work
protein expr as reg
takes longer, more long term solu, requires changes to transc/l, mRNA stability, etc
covalent mods
requires separate e to put on and take off, not permanent, can inc/dec activity or turn on/off e (dept on e)
these are groups like me or phosphates that aren’t proteins like am and require e
can direct change in activity themselves or make a binding site for something else to bind and change e activity (direct, indirect) - can be combo of covalent mod and am
common covalent mods
phosphorylation - adding phosphate
adenytlation - add adenine
methylation - add methyl groups
phosphorylation
to tyr, ser, thr, his residues
[s name] kinase puts on
[s name] phosphatase takes off
adenylation
takes adenine from ATP, adds it to tyr residues
methylation
adds me groups to glu residues
localization
cell sequesters active e in region w/o s so when the cell is in environ w/ s, it releases e into cytoplasm
spatial/environ dept reg