Viral genomes and diversity
1/39
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
40 Terms
What is a virion?
A complete infectious viral particle consisting of a nucleic acid genome surrounded by a protein coat, and sometimes a lipid envelope
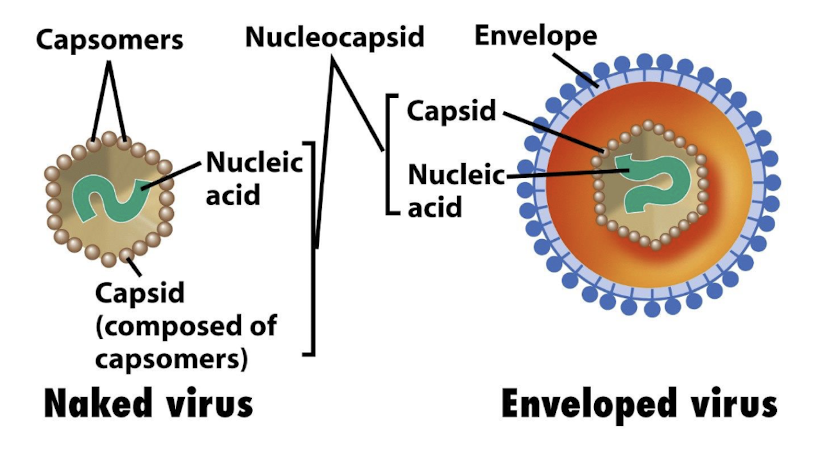
What is a capsid?
The protein shell that protects viral nucleic acid; may have icosahedral, helical, or complex morphology

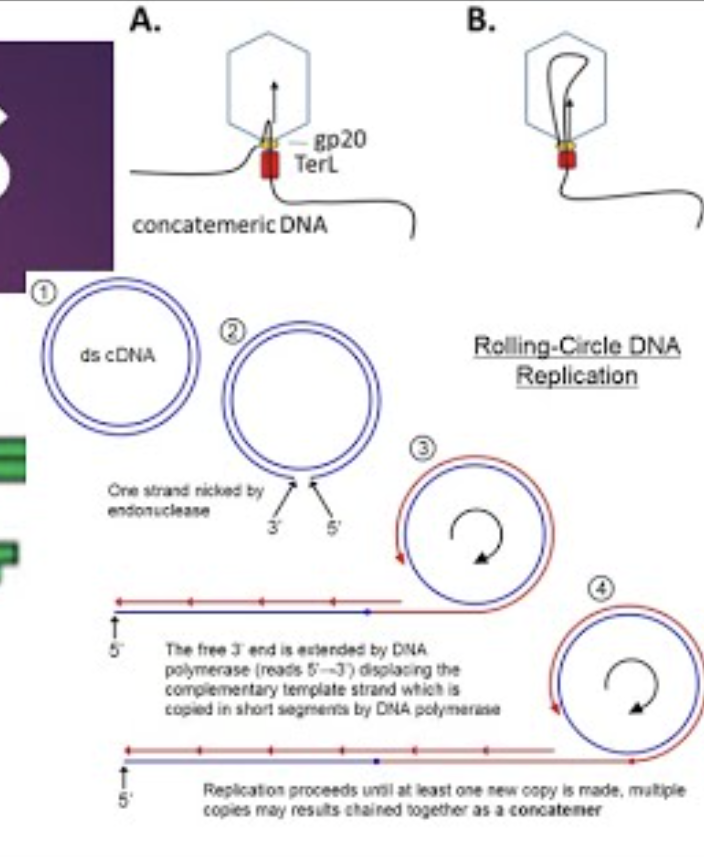
What is a concatamer?
A long, continuous DNA strand containing multiple genome copies produced during rolling-circle replication
What is a provirus?
A viral genome integrated into host DNA (e.g., HIV). It is replicated along with the host genome and may be latent.
What is antigenic variation?
The ability of a pathogen to alter surface antigens to evade immune detection (e.g., influenza antigenic shift; trypanosomes)
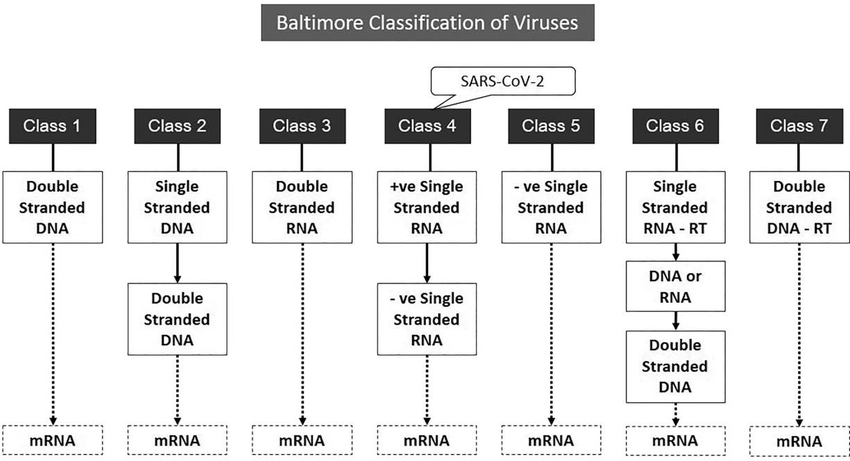
What is the Baltimore classification scheme?
A system that classifies viruses based on their genome type and the pathway used to generate mRNA for protein synthesis.
It includes 7 classes (I–VII) depending on whether genomes are DNA or RNA, single- or double-stranded, and positive or negative sense.
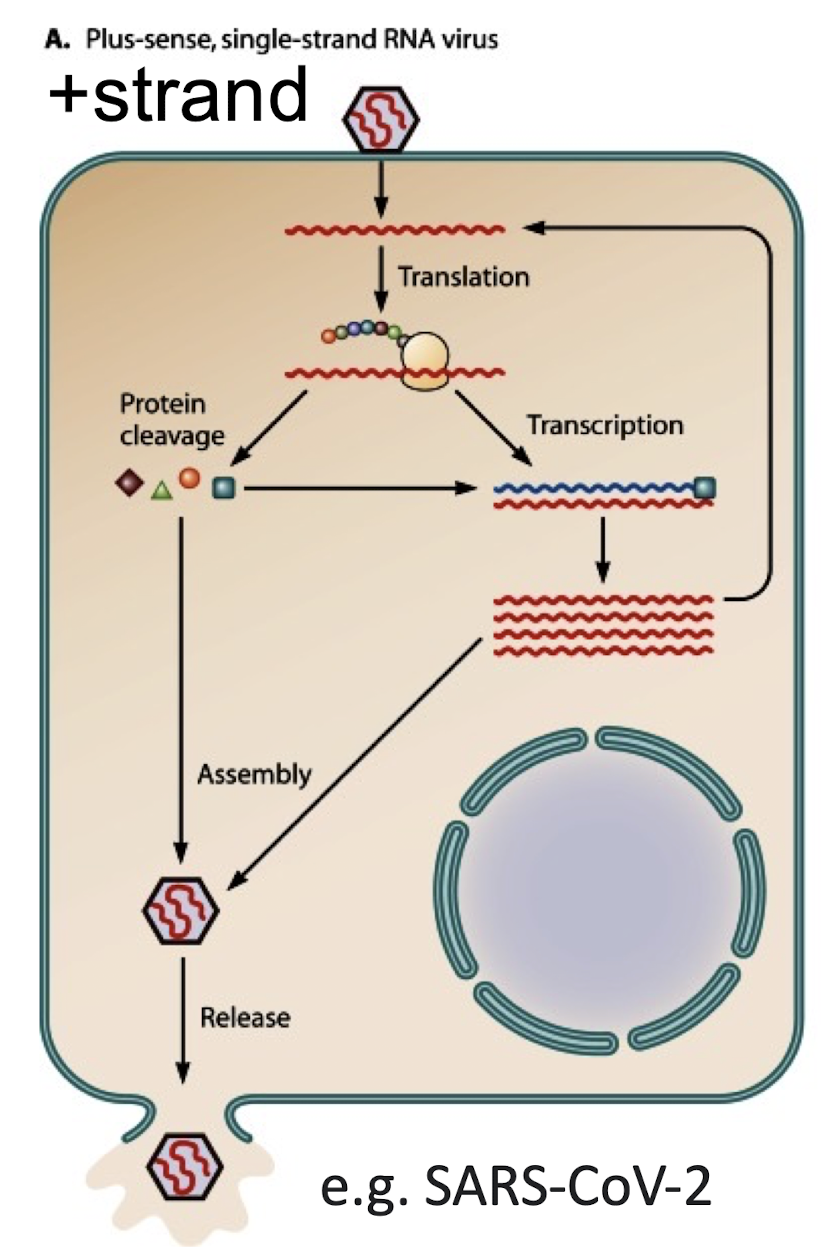
What is a positive-sense (+) RNA genome?
A type of single-stranded RNA virus whose RNA CAN be directly translated into proteins.
eg SARS-CoV-2
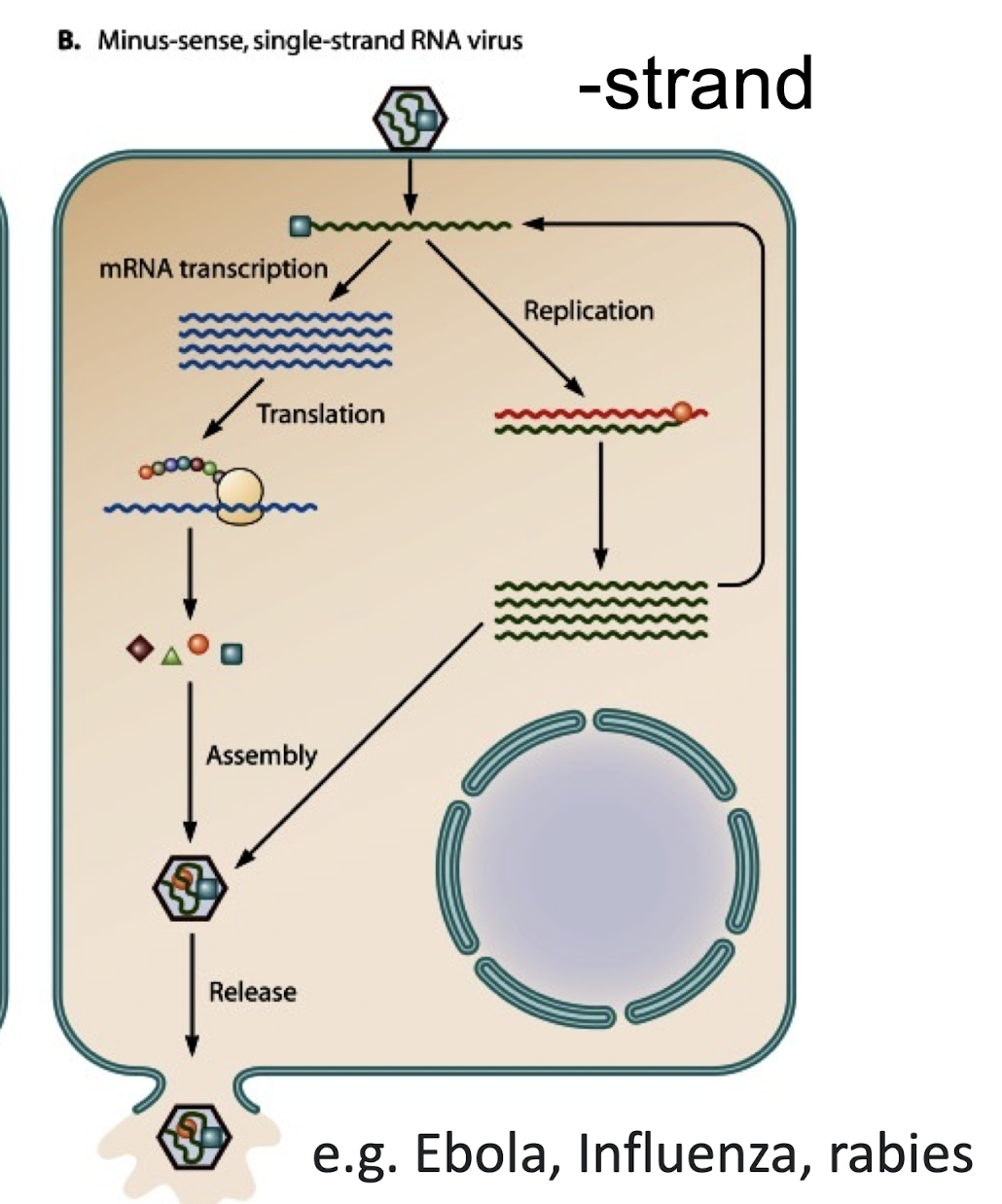
What is a negative-sense (–) RNA genome?
An RNA genome complementary to viral mRNA; it must be copied by viral RNA-dependent RNA polymerase into a positive-sense strand before translation.
What is rolling-circle replication and where does it occur?
A replication mechanism in which a circular DNA/RNA template produces long concatameric strands(contains multiple copies of the same DNA sequence linked in series) that are later cleaved into unit genomes.
Occurs in Class II ssDNA phages (e.g., M13, ΦX174)
Class I viruses (herpesviruses, λ phage)
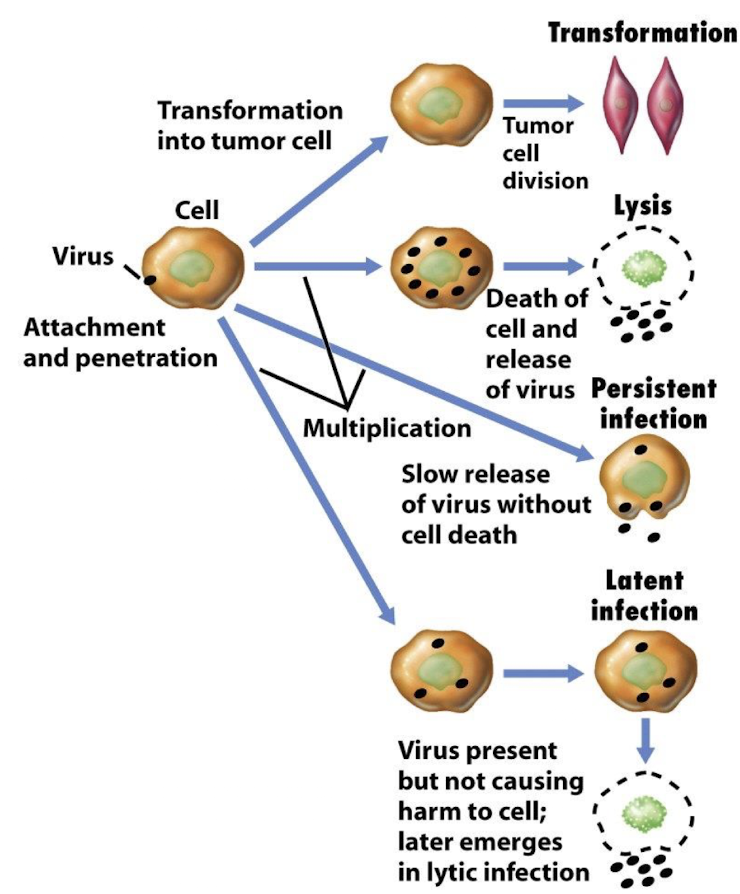
What are lytic, persistent, and latent infections?
Lytic: Cell is destroyed upon virus release
Persistent: Slow, continuous release without immediate cell death.
Latent: Viral genome remains silent, may reactivate later (e.g., herpes).
Some viruses can transform host cells into tumours e.g. Polyoma viruses in non-permissive cells cause viral DNA to get integrated into the host chromosome + cause a tumour
How do Class I dsDNA viruses generate mRNA?
Use host (or viral) DNA-dependent RNA polymerase to transcribe mRNA from dsDNA. Their replication resembles that of host cells.
e.g. many bacteriophages, Pox viruses
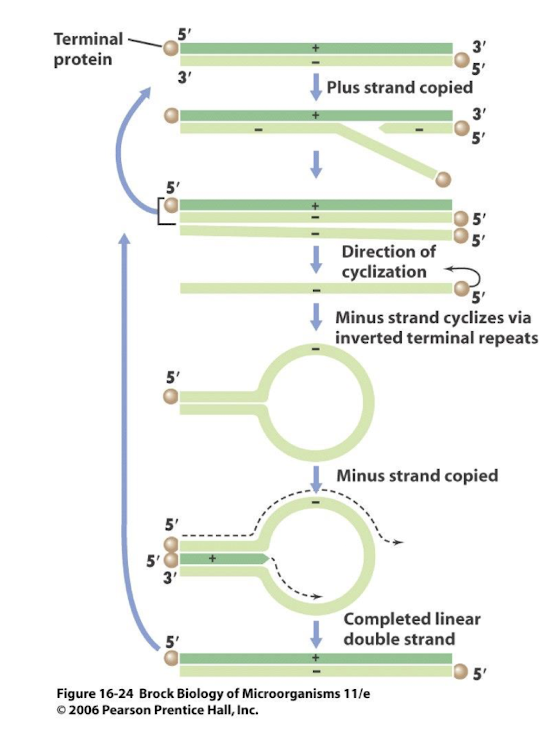
How do Class I viruses replicate their genomes and what is an exception?
Replication usually occurs in the nucleus using host DNA polymerases, except poxviruses, which replicate in the cytoplasm and encode their own polymerases.
Example replication mechanism of a Class I virus?
Herpesvirus uses rolling-circle replication after early/late gene expression phases.
Pox virus: include variola major, vaccinia, and cowpox virus.
What are traits of Pox viruses (Class I)?
Large and brick shaped virions
Replicate in the cytoplasm
Virus encodes its own DNA and RNA polymerases
Vaccination with vaccinia virus led to eradication of small pox
What is special about adenovirus genome replication?
It is conservative, not semiconservative; the negative sense single strands are synthesised 1st, then converted to dsDNA.
How do Class II ssDNA viruses generate mRNA?
The ssDNA is first converted into a dsDNA replicative form by host enzymes, which is then transcribed to mRNA
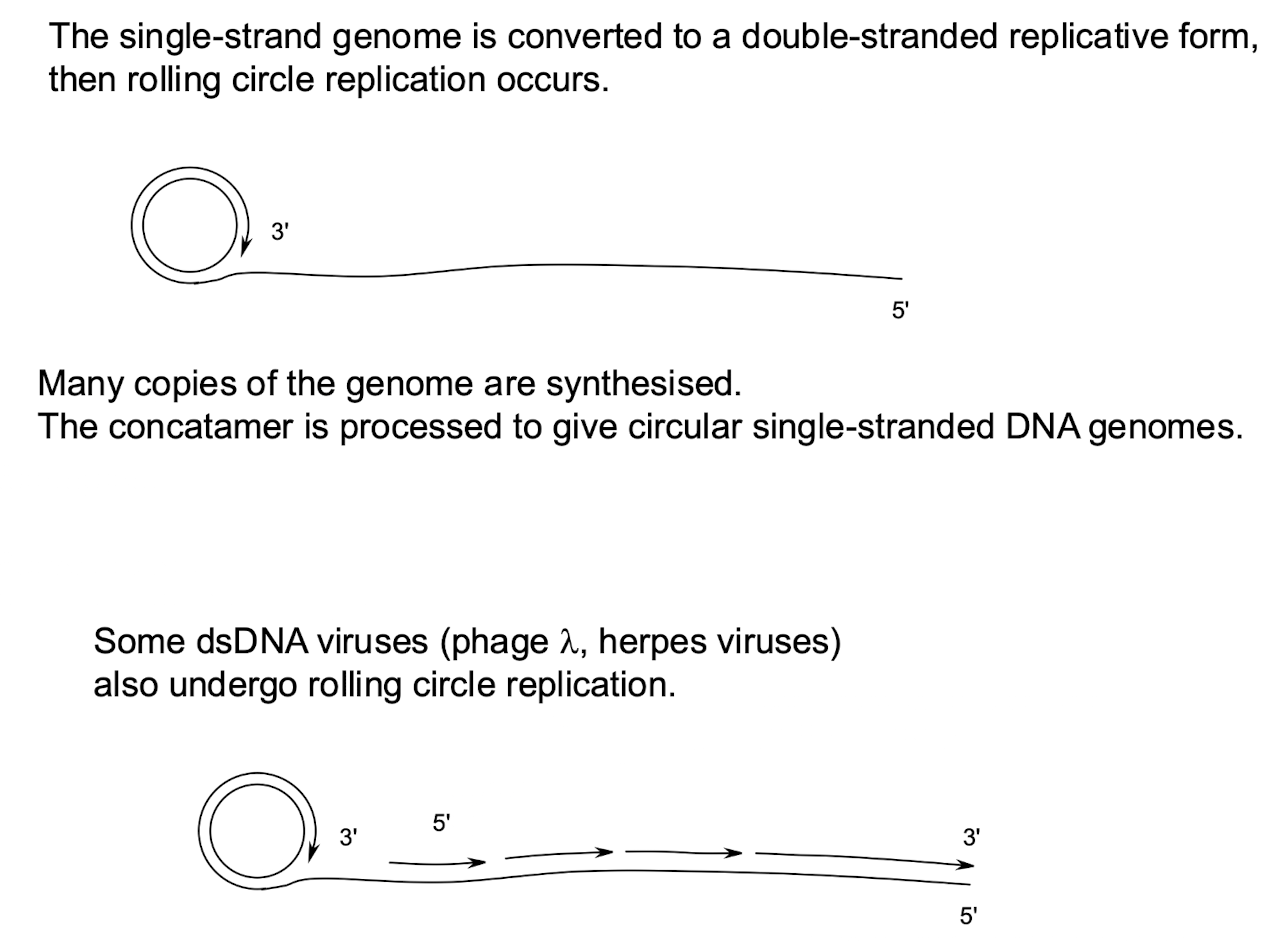
How do Class II viruses replicate genomes?
Through rolling-circle replication, producing concatamers later processed to give single-stranded DNA genomes.
e.g. Parvoviruses, filamentous M13, and ΦX174 bacteriophages
What is an example of a Class III dsRNA virus?
Reoviruses (Respiratory and enteric orphan viruses)
e.g. Rotavirus which causes infant diarrhoea.
Icosahedral nucleocapsid with two protein coats.
Genome consists of 10 to 12 linear ds RNA molecules totalling 18 – 30 kb in size
How do Class III (+ sense) dsRNA viruses generate mRNA?
e.g. Reovirus
Viral RNA-dependent RNA polymerase (RNA replicase) within the virion synthesizes positive-sense mRNAs from each dsRNA segment
lacks proof-reading ability therefore leads to high mutation rates.
What is an RNA-dependent RNA polymerase (replicase)?
A viral enzyme that synthesises RNA from an RNA template; essential for Classes III, IV (genome replication), and V.
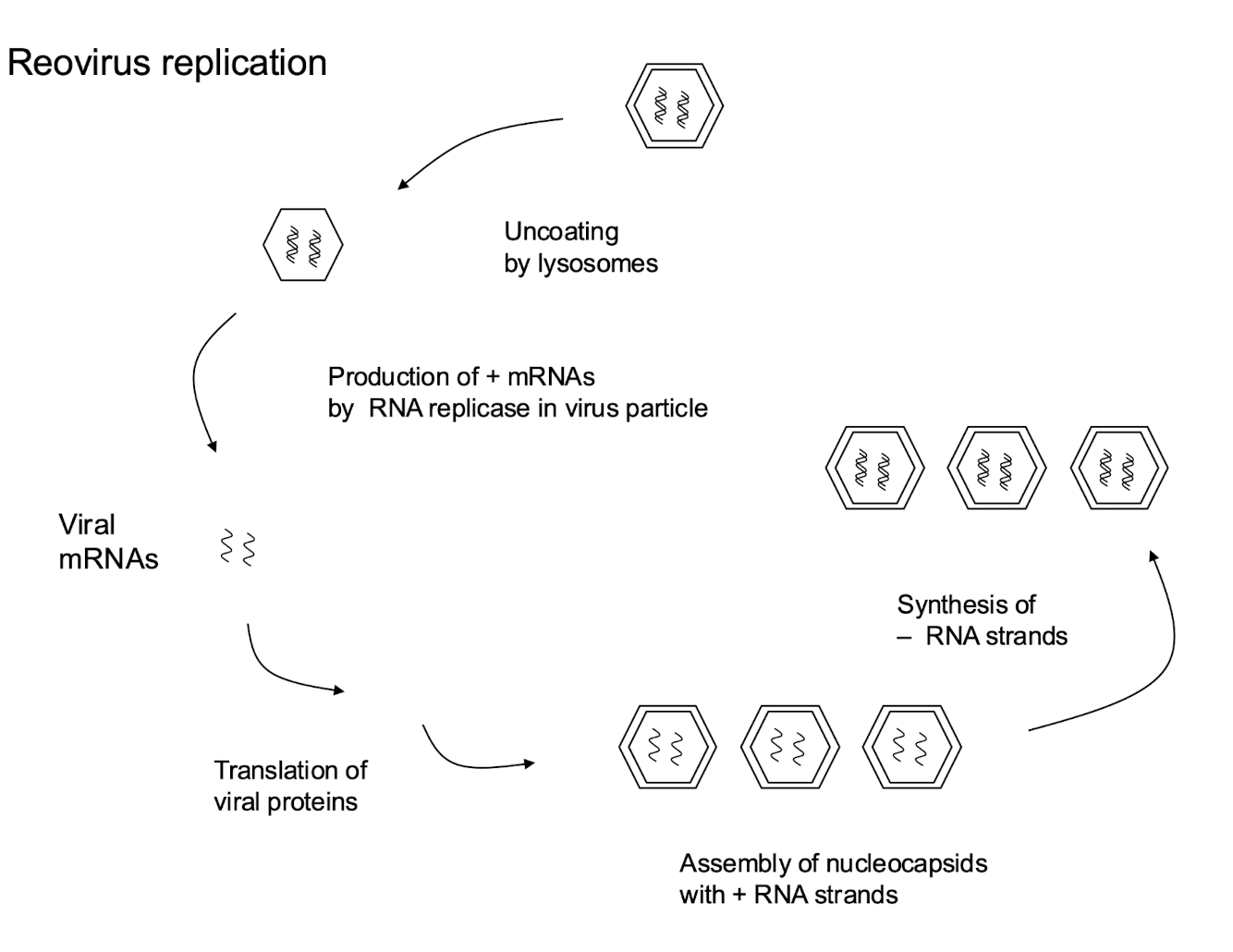
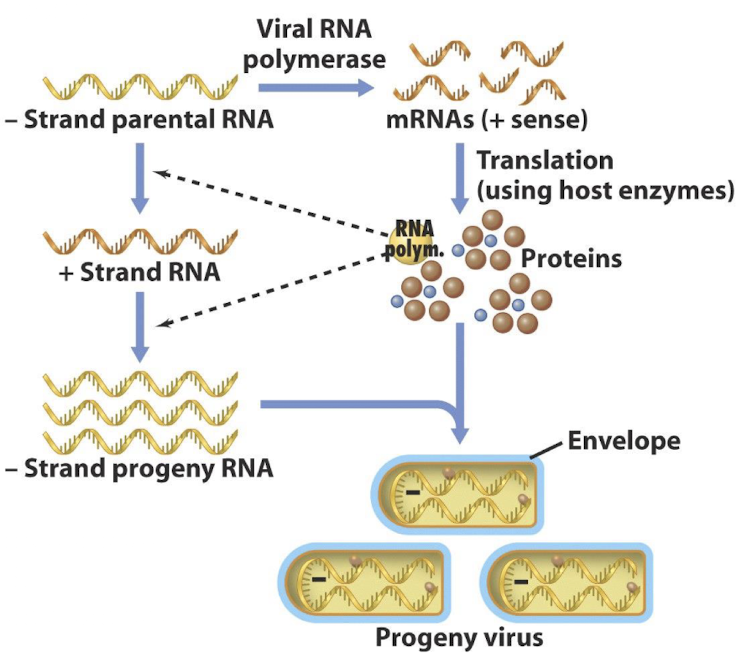
How do Class III viruses replicate their genomes?
Virus is uncoated by lysosomes
(+) mRNAs are made by RNA replicase in virus particle
After producing ( + ) mRNA strands, these serve as templates for the translation of viral proteins
The assembly of nucleocapsids with (+) RNA strands leads to synthesis of new ( – ) RNA strands, forming new dsRNA segments
How do Class IV viruses express their genes?
Their genomes act directly as mRNA, translated immediately upon entry into the host cell.
These positive-sense ssRNA genomes can be directly translated by the host's ribosomes to produce viral proteins.
How do Class IV viruses e.g. Poliovirus replicate genomes?
The VpG protein binds to 5 ′ ends of RNA.
The 3 ′ end has a polyA tail.
The genome acts as an mRNA initially.
Translation gives a polyprotein precursor of structural proteins, a protease, and RNA replicase.
Minus strands are made and act as templates for production of new genomes
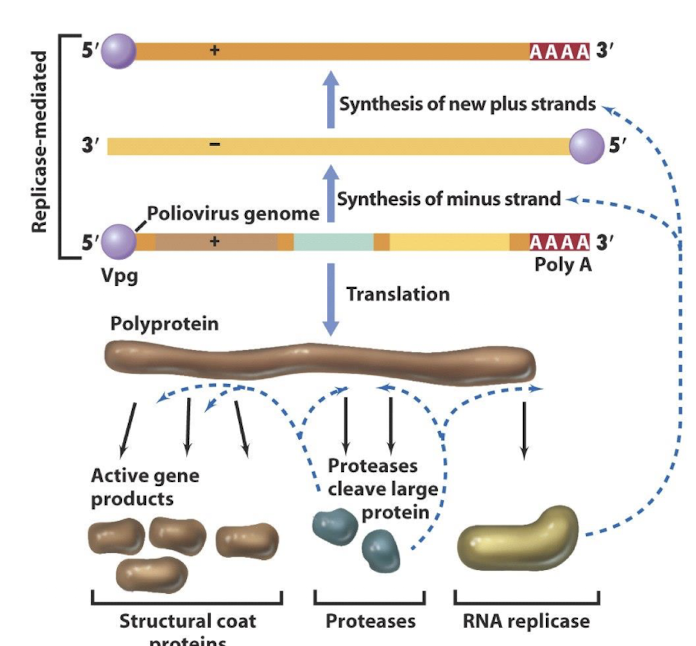
Example life cycle feature of poliovirus?
Produces a single polyprotein that is autoproteolytically cleaved into functional proteins.
VPg on the 5′ end and poly-A tail on 3′ end are key features of its genomic structure.
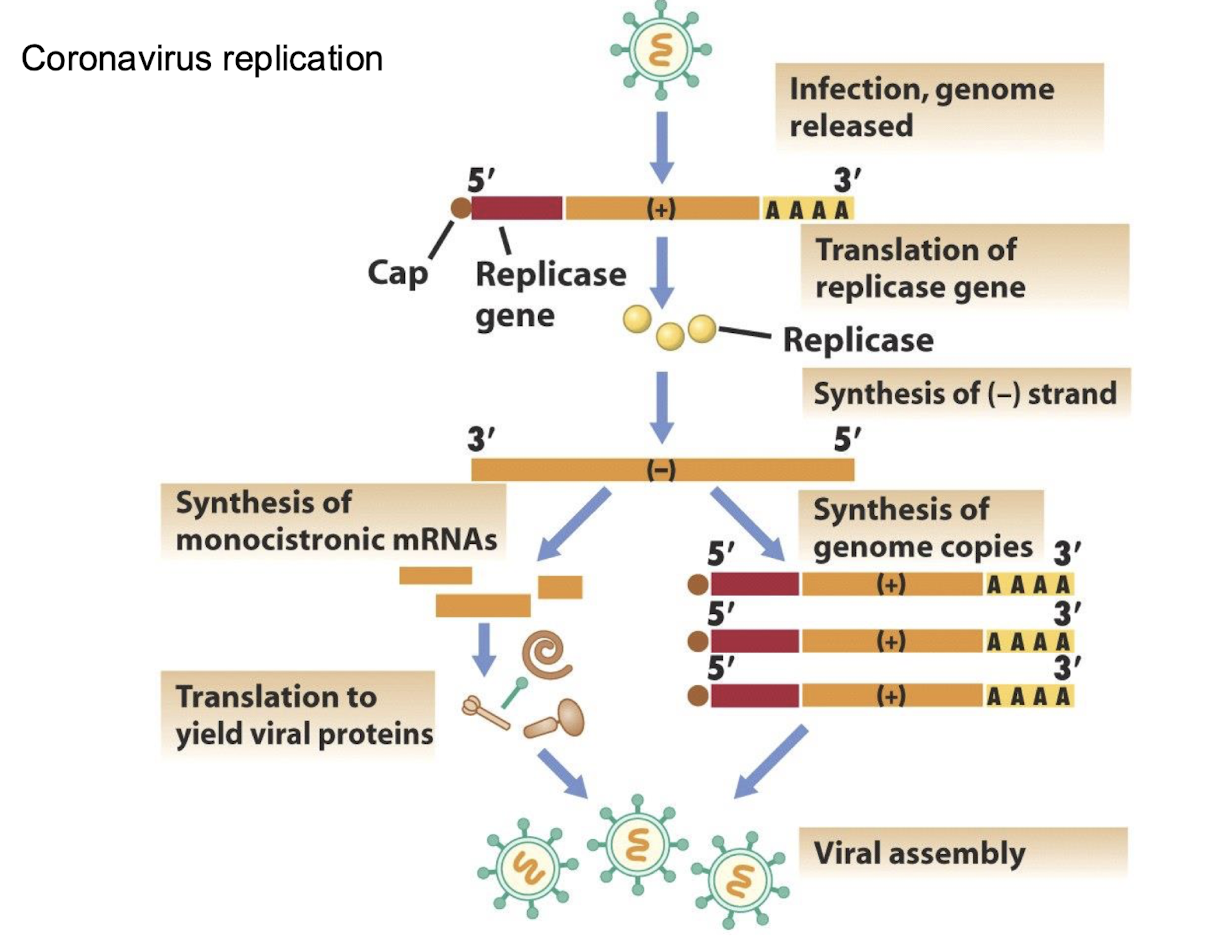
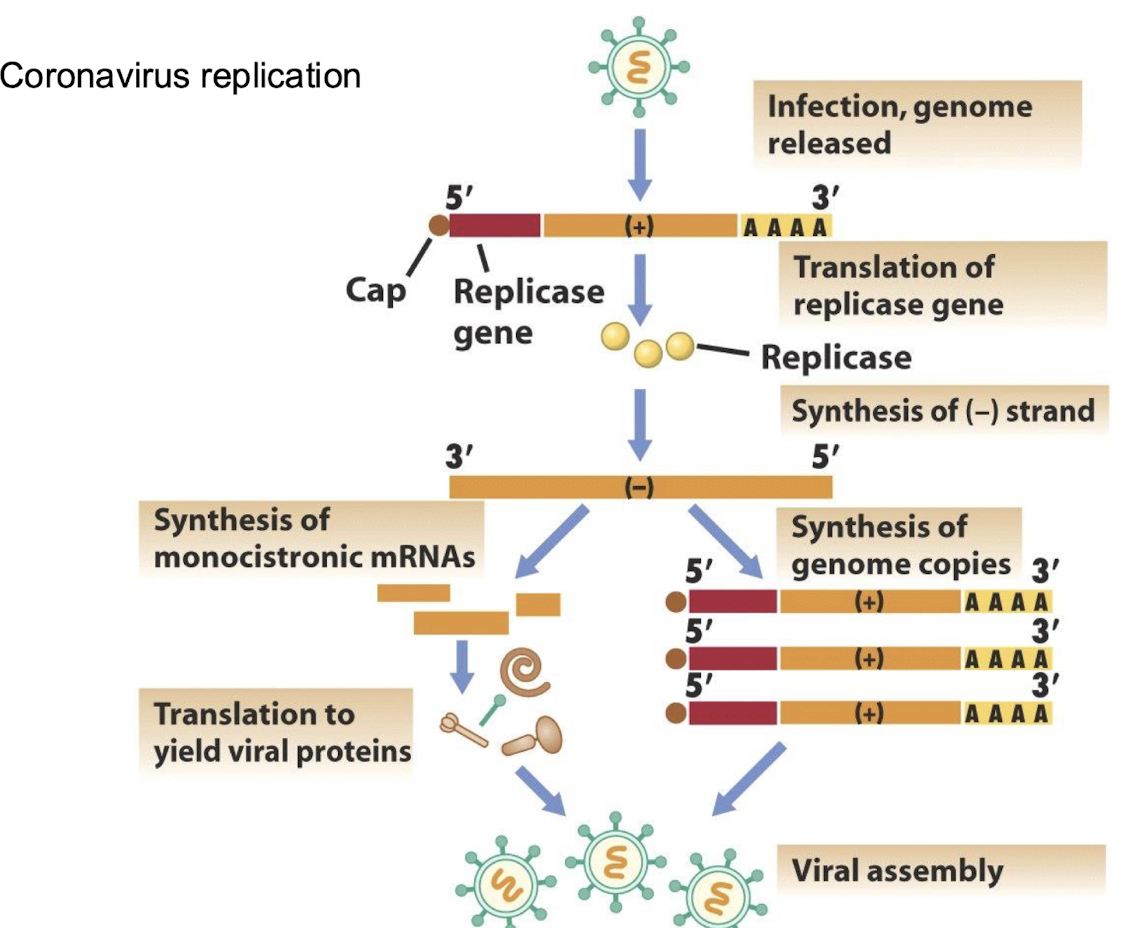
How do coronaviruses express multiple proteins from a +RNA genome?
Through a nested set of monocistronic subgenomic mRNAs, each translated into specific proteins that are essential for viral replication and assembly.
How do Class V (Negative-strand ssRNA) viruses express genes?
They must carry a viral RNA-dependent RNA polymerase in the virion to convert (–) RNA into ( + ) mRNAs
e.g. Rabies, Influenza and Ebola viruses.
What is antigenic shift in influenza?
A major change in the virus's surface proteins due to the reassortment of genome segments when two different strains infect the same cell, leading to new subtypes that can evade immunity.
Key glycoproteins in influenza and their roles?
Haemagglutinin (HA): mediates attachment; immunogenic protein in flu vaccines
Neuraminidase (NA): cleaves sialic acid, enabling viral release from host cell.
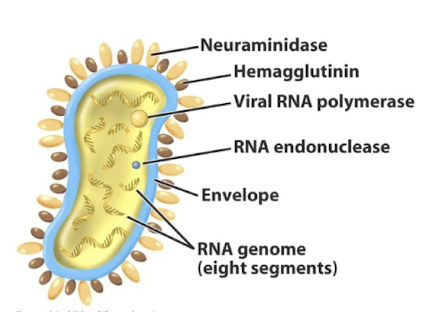
What is antigenic shift?
The ability of a pathogen to alter surface antigens / combine genome segments from 2 different viruses to evade immune detection (e.g., influenza antigenic shift; trypanosomes)
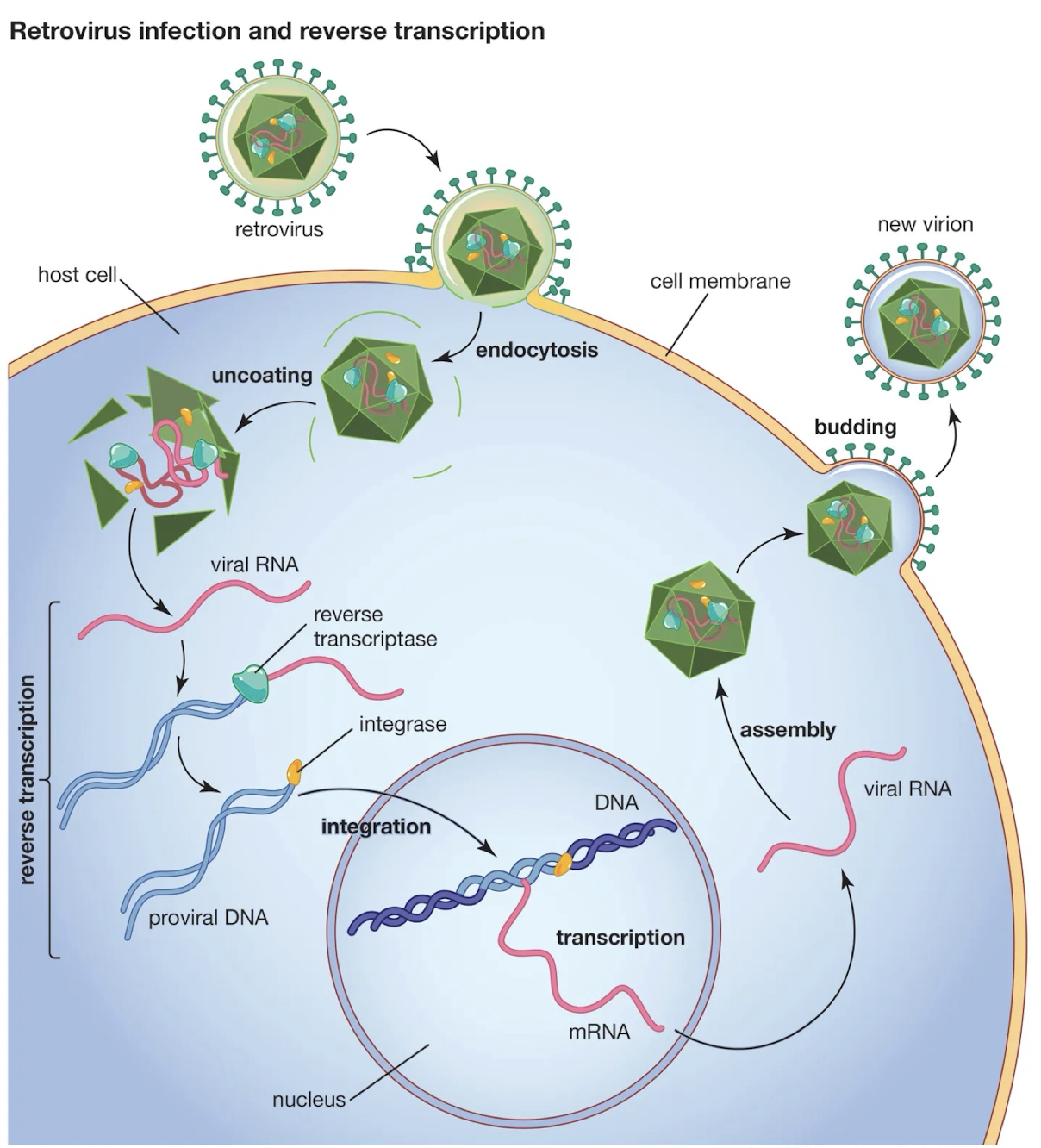
What is reverse transcriptase?
A viral enzyme that synthesises DNA from an RNA template; used by Classes VI (retroviruses) and VII (Hepadnaviruses).
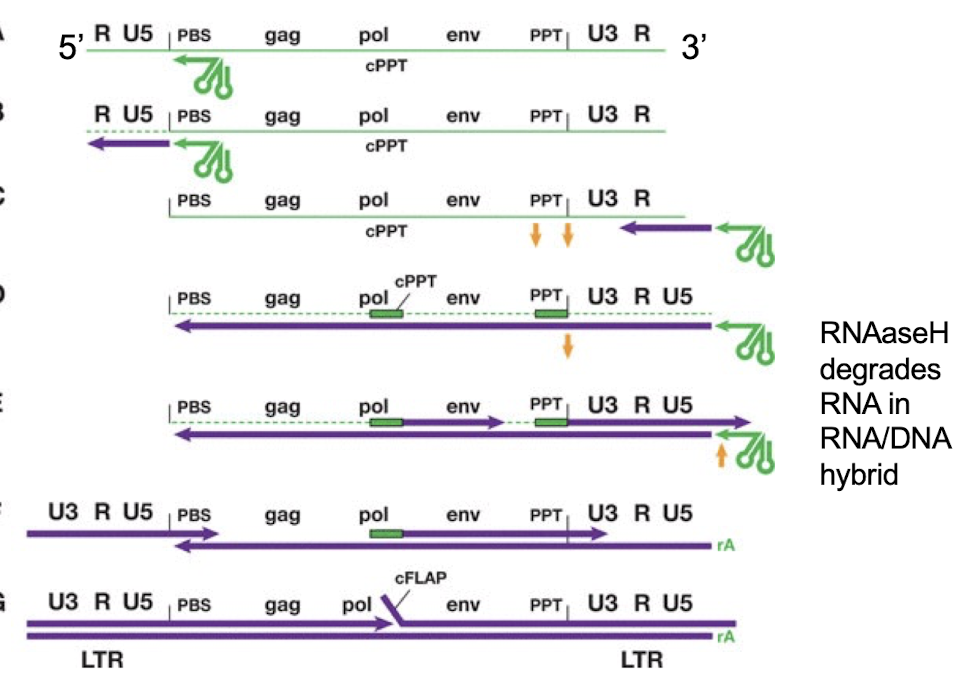
_________ is a specific region on the RNA genome where a _________ binds to initiate reverse transcription
Primer Binding Site (PBS) , tRNA primer
How do Class VI viruses (retroviruses) generate mRNA?
After reverse transcription of the RNA genome into dsDNA, the integrated provirus is transcribed by host RNAPol II to make viral mRNAs.
What is a key example of Class VI viruses and how is the genome replicated?
HIV (Human Immunodeficiency Virus)
The RNA in the virion is positive strand.
After infection, it is copied into DNA by reverse transcriptase.
Eventually a dsDNA version of the viral genome is formed and this integrates into a host chromosome.
Integration can occur anywhere in the genome but usually occurs within an area of active chromatin.
The integrated copy remains latent as a provirus but it can be induced and transcribed by the cellular RNA Pol to make new viral genomes as well as mRNAs that are translated to make viral particle proteins
____________ are repeated sequences at the ends of the double-stranded DNA generated from an RNA genome, resulting from the ___________
long terminal repeats (LTRs), reverse transcription
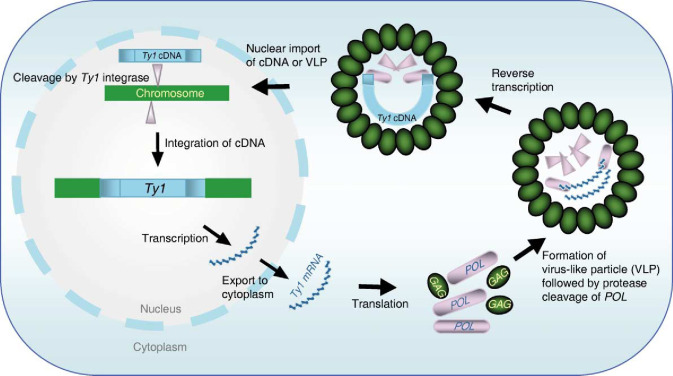
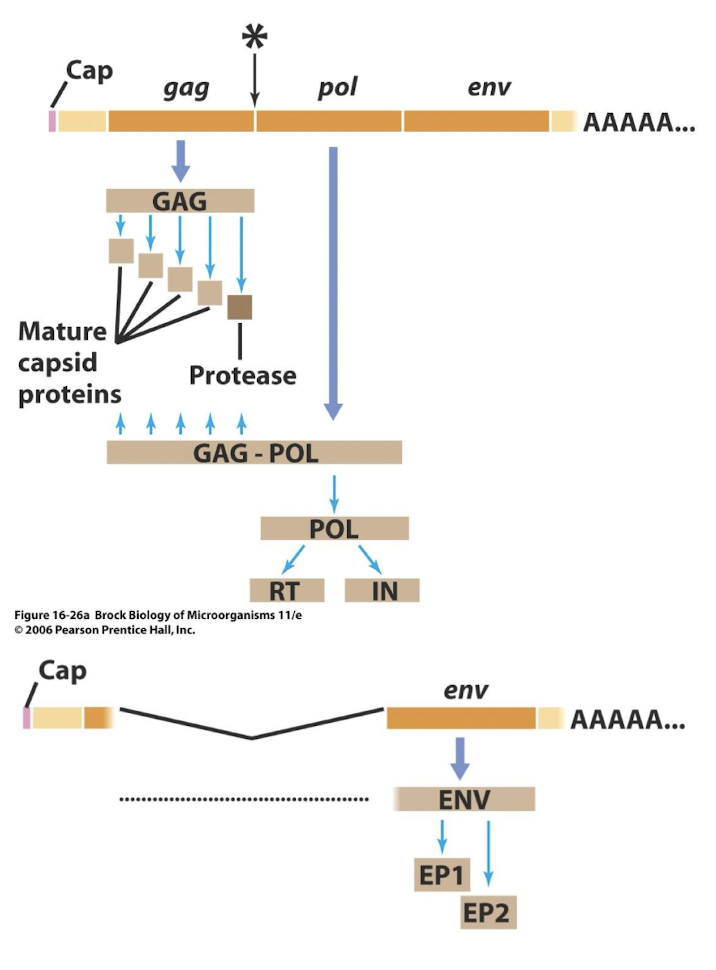
What proteins are produced from the gag–pol–env reading frame in HIV?
Frameshift suppression allows translation of Pol - precursors of RT, integrase
Late mRNA splicing produces envelope (Env) glycoproteins and structural proteins (gag) that form the viral core.
How do Class VII viruses (dsDNA viruses requiring an RNA intermediate) generate mRNA?
Their partially dsDNA genome becomes fully ds + circular on infection
Host RNA polymerase II transcribes mRNAs and an RNA copy of the genome
Reverse transcriptase copies the pregenomic RNA into partially dsDNA
Maturing virus particles are exported through the ER and Golgi
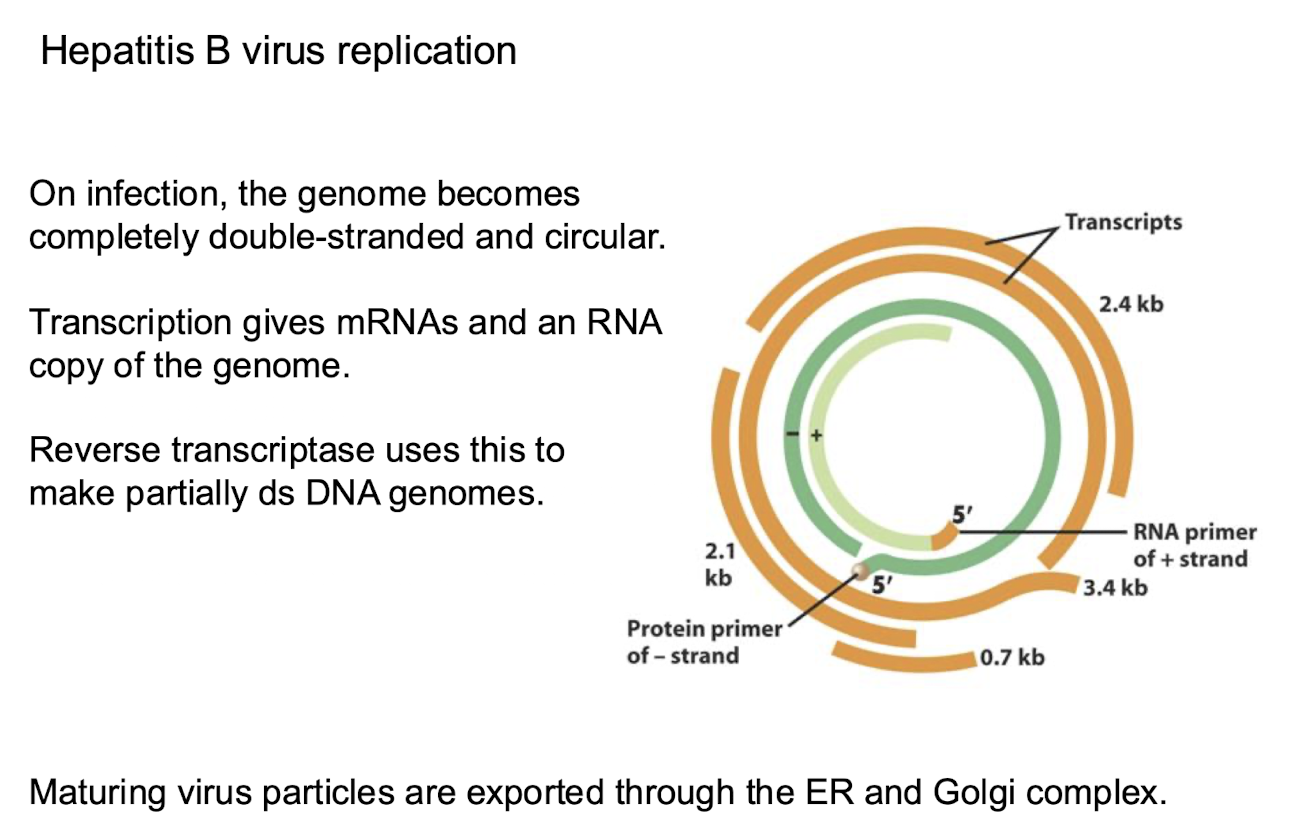
Example of Class VII virus?
Hepatitis B virus (HBV), with 3–4 kb partially double-stranded DNA genome
What are examples of subviral agents?
Viroids
Infectious RNA molecules
small circular ssRNAs
Only infect plants
Plants have enzymes that replicate RNA from an RNA template.
Catalytic RNA can excise themselves from a concatameric precursor.
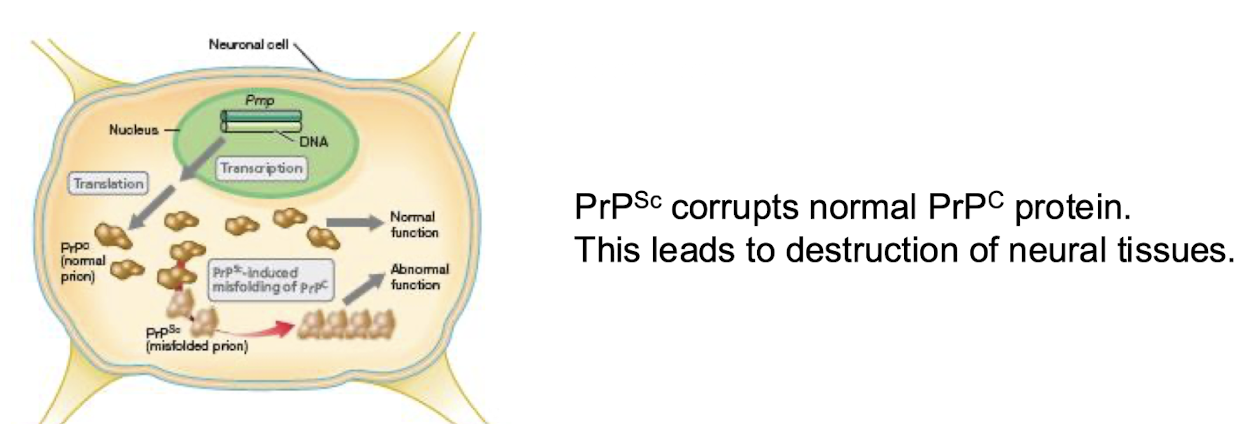
What are prions?
Infectious agents composed only of protein.
Cause TSEs, scrapie in sheep, BSE in cattle, CJD in humans.
The normal cellular protein PrpC has mostly a-helical structure.
The pathogenic form PrpSc has mostly b-sheet + corrupts the normal form, + disrupts brain function.
How should I structure an essay on the Baltimore classification and replication strategies?
Intro: Define viruses, intracellular parasitism, and the logic of the Baltimore scheme.
Class-by-class structure:
– Genome type
– How mRNA is generated
– Mechanism of genome replication
– Key examples and special featuresLife cycles: Lytic/persistent/latent patterns and integration (Class VI, VII, herpes).
Conclusion: Emphasise how genome type dictates replication strategy and pathogenic outcomes.