Week 10: DNA Damage, Repair + Genome Editing
1/33
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
34 Terms
DNA damage
chemical changes to nitrogen rich bases
N-rich bases not inert → reactive
can lead to changes in BP potential → mutations
can lead to double-strand breaks → genome instability
can be repaired or cells undergo cell death (apoptosis)
caused by
oxidative damage
spontaneous deamination
loss of base
replication errors
UV exposure = photo-cross-linking
chemical exposure = alkylation/methylation damage
spontaneous deamination
cytosine loses exocyclic amino group → turns into uracil
uracil chemically similar to thymidine = can base pair w/ adenine
leads to AT mutation if unrepaired
recognized as foreign in DNA + removed
why DNA has thymine rather than uracil
other bases can also undergo reaction
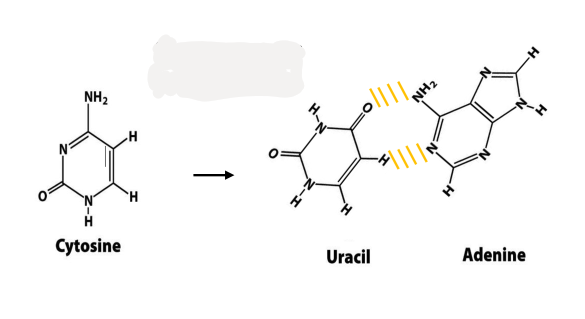
cytosine deamination
turns into uracil
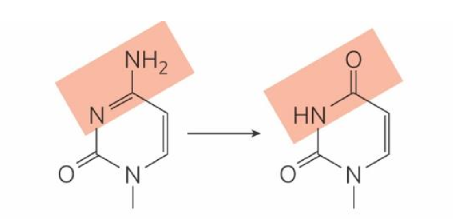
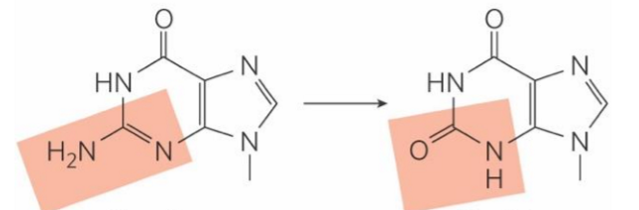
adenine deamination
turns into hypoxanthine
hypoxanthine = non-coding
recognized by cell as damage → removed
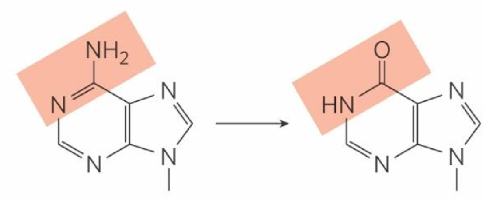
5-methylcytosine deamination
turns into thymine
_______ = modified base in DNA w/ high frequency in patches
enhances/represses transcription = controller
cell can’t tell that base is incorrect → won’t remove
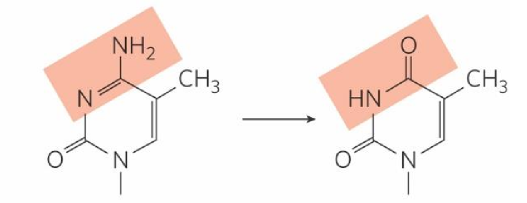
guanine deamination
turns into xanthine
xanthine
recognized by cell as damage → removed
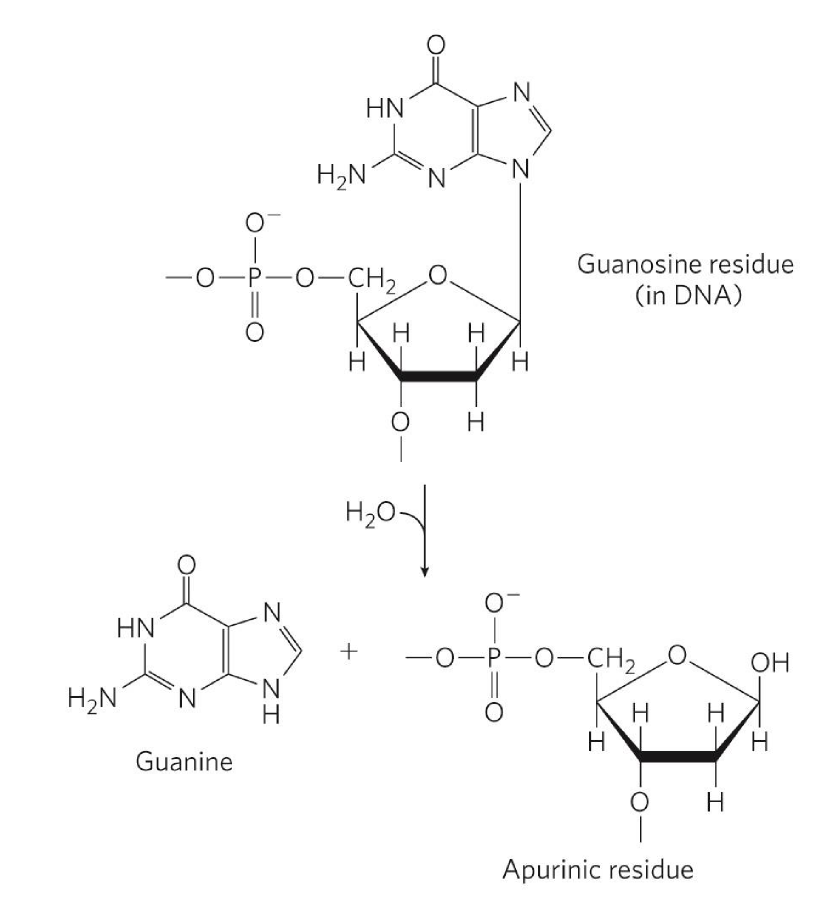
depurination
hydrolysis of N-B-glyosyl bond b/w base + pentose
H2O comes in + hydrolyzes bond = base floats away
phosphodiester bond still stuck in DNA
creates AP (apurinic/apyrimidic) site = abasic
info void region generated
more common w/ purine
double-rings = good-leaving group
why AMP is used as activator
oxidative damage
reactive oxygen species damage DNA
mitochondria = oxidative compartment → generates lots of energy via oxidative phosphorylation
hydroxyl (OH) radicals responsible for most damage
cells have defense system to destroy reactive oxygen species
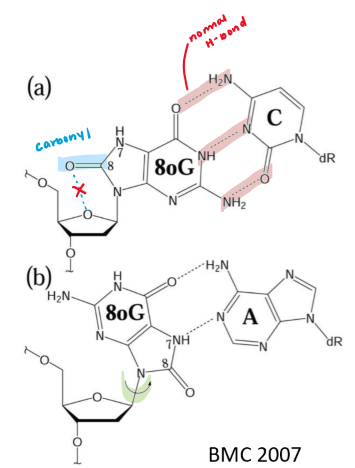
eg. guanine → 8-oxo-guanine
anti-conformation usually favoured
new steric clash b/w carbonyl + ribose oxygen = syn-conformation favoured
⬆ Hoogsteen base-pairing b/w G + A
after replication → A-T pair
endogenous damage
spontaneous
deamination
changes H-bonds → drives mutations after replication
loss of base
generates abasic sites → loss of info
oxidative damage
changes from anti → syn-conformation
changes base pairing
exogenous damage
DNA damage caused by external factors such as chemicals, radiation, or infectious agents
photo-cross linking from UV radiation
covalent bonds form b/w bases on same strand instead of H-bonds w/ base across → lesion
alkylation from chemical exposure
addition of alkyl (eg. methyl) groups to base → affects base pairing
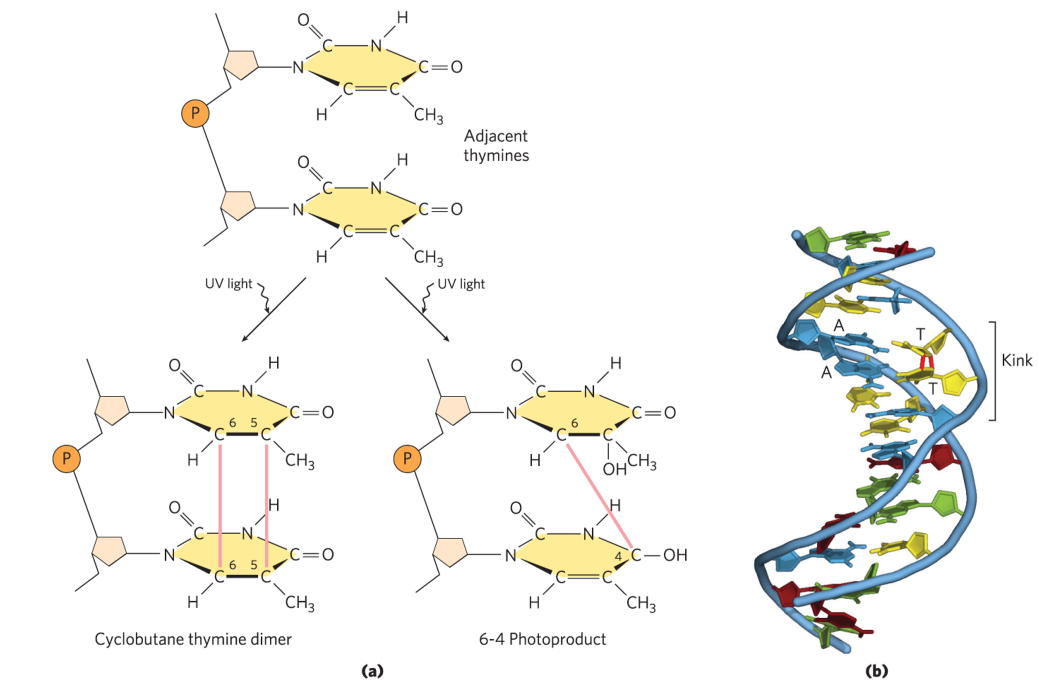
photo-cross-linking
caused by UV damage or radiation
UV
cyclobutane pyrimidine dimers
6-4 photoproduct
ionizing radiation
ring opening
base fragmentation
breaks in covalent backbone of nucleic acids
causes adjacent pyrimidine bases to bond with each other, forming dimers
instead of making H-bonds w/ something across → makes covalent bonds w/ itself
squishes 2 bases together + prevents info from being accessed
alters DNA structure + stalls replication → polymerase can’t read + falls off
nEta (translesion polymerase)
bypasses damage from cross-links
no editing site
not reading template
larger active site fits distorted cross-linked Ts
adds 2 dAs to UV damage to synthesis strand
falls off → not processing
enables Pol III (prokaryotes) or Pol δ/ε (eukaryotes) access to 3’OH
can take over + replicate
damage still there
usually adds dC if 8-oxo-guanine
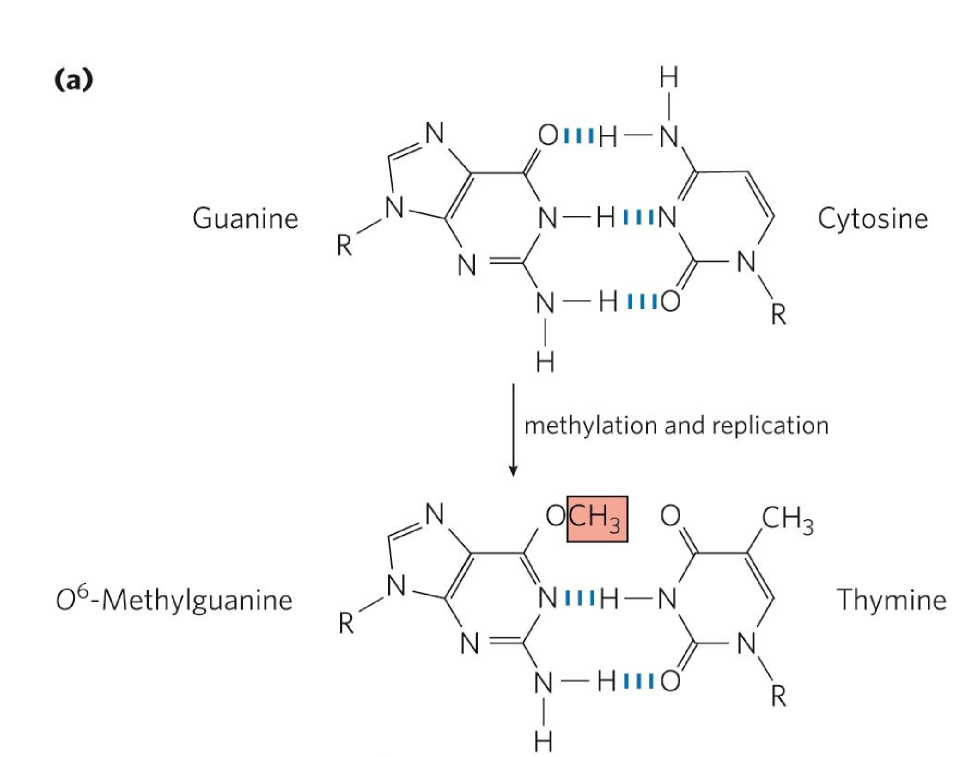
alkylation damage
addition of alkyl groups to DNA bases
from chemicals, eg. cigarette smoke, mold, burnt foods
leads to mutations and disruption of DNA replication
ex. O6-methylguanine → methyl added to gunanine carbonyl group
common + highly mutagenic lesion
methyl group blocks repulsive interaction b/w G + T oxygens
enables pairing w/ thymine rather than cytosine
DNA repair hypothesis
DNA = only biological molecule that life takes time + effort to repair
DNA stability essential for encoding proteins/RNA
DNA easily damaged
mutation
permanent change in NT sequence
1+ mutation → can become cancer
types:
substitution = replacement of BP
insertion = addition of 1+ BP
deletion = deletion of 1+ BP
caused by:
polymerase errors
DNA damage that then favours new BP interaction
silent mutation
mutation that affects nonessential DNA or has neglible effect on gene function
Ames test
test mutagenic potential of chemical compounds by observing if they cause mutations in DNA of bacteria
Salmonella typhimurium w/ mutation in histidine synthesis pathway grown on histidine-free plates
something blocking histidine synthesis
Whatman filter paper soaked in potential mutagen
colonies arise in mutagenic conditions
mutation relieves histidine synthesis blockage
DNA repair mechanisms
base modification, abasic site
base excision repair (BER)
crosslinks
nucleotide excision repair (NER)
mismatches, insertions, deletions
mismatch repair (MMR)
double strand breaks (DSBs)
homologous recombination (HR)
non-homologous end joining (NHEJ)
Dam methylase
functions in mismatch repair in E.coli - methylates GATC sites on adenine residues
use single-carbon transfer co-factor SAM
~1min after replication, ______ methylates sites in newly synthesized daughter strand
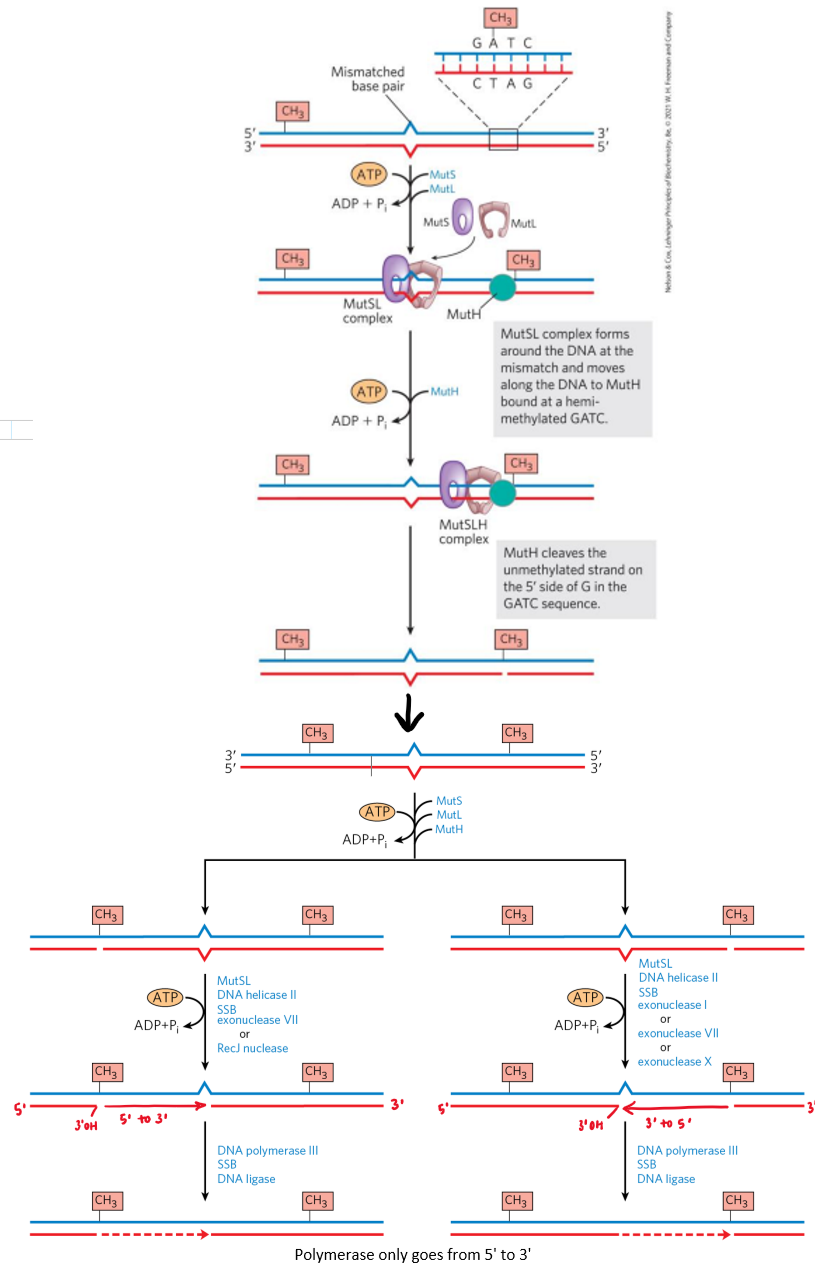
mismatch repair (MMR)
fixes base-base mispairing + insertion/deletion
identification
Dam methylase recognizes GATC site on parent strand + methylates
reads site in 5’→3’ direction
shortly after replication occurs → daughter strand doesn’t have methylation
know which strand is parent + where mistake occurred
MutS + MutL loaded onto dsDNA at lesion (mismtach) using ATP
complex slides along DNA until GATC methylation site encountered
can slide right or left → direction-independent
MutH cleaves non-methylated strand = single-strand nick made
repair
helicase unwinds DNA at nick site + moves to lesion
NTs b/w nick + lesion removed by exonuclease
can be 5’→3’ or 3’→5’
nick or exonuclease generates free 3’OH
non-damaged strand used as repair template for DNA polymerase
gap at end of repair sealed w/ ligase
base excision repair (BER)
fixes modified bases + abasic sites
damaged base recognized by specific glycosylase → base removed = abasic site made
endonuclease cleaves phosphodiester backbone at basic site → 5’ phosphate nick
DNA pol I replaces missing base + does short extension via nick translation
gap filled by DNA ligase
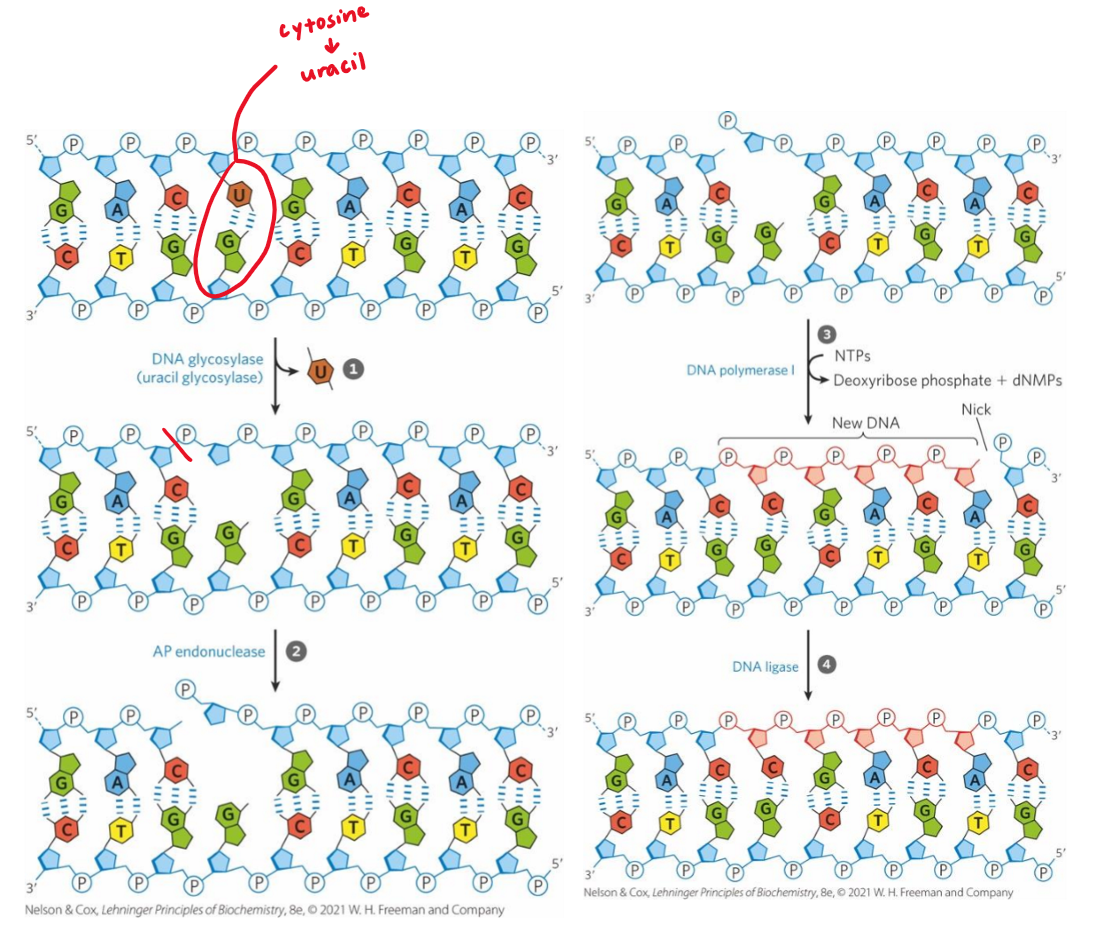
DNA glycosylases
recognize common DNA lesions + remove affected bases by cleaving N-glycosyl bond in BER → abasic site made
generally specific for 1 lesion type
eg. pocket inside specific for uracil
if uracil present in DNA → fit into pocket
senses it exists → shouldn’t be in DNA
hydrolyzes N-glycosidic bond
abasic site generated
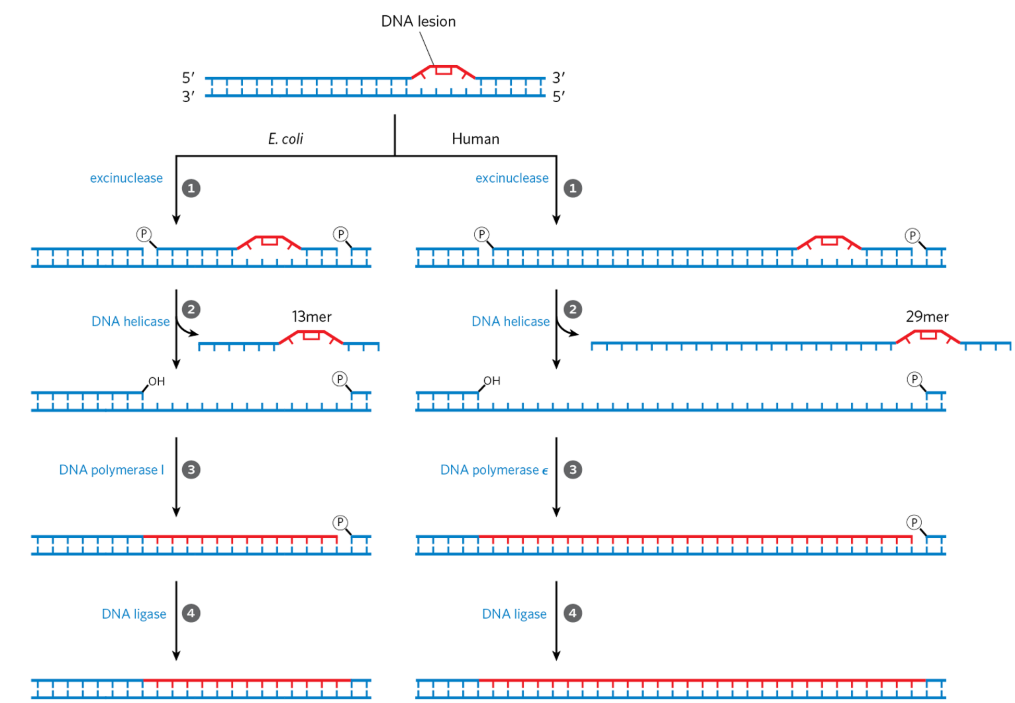
nucleotide excision repair (NER)
repairs DNA lesions that cause large distortions in helical structure, ie. photo-crosslinks
lesion attracts excinuclease complex
excinuclease: multisubunit enzyme that hydrolyzes 2 phosphodiester bonds on either side of distortion
proteins come along + recognize lesion
stay on strand + move along
make nick in backbone on either side of lesion
larger fragment stuck on template via base-stacking + H-bonds
helicase zips off fragment from template
3’OH on single-stranded template = substrate for DNA pol
DNA pol I (E.coli) or DNA pol ε (humans) fill gap
DNA ligase seals nick
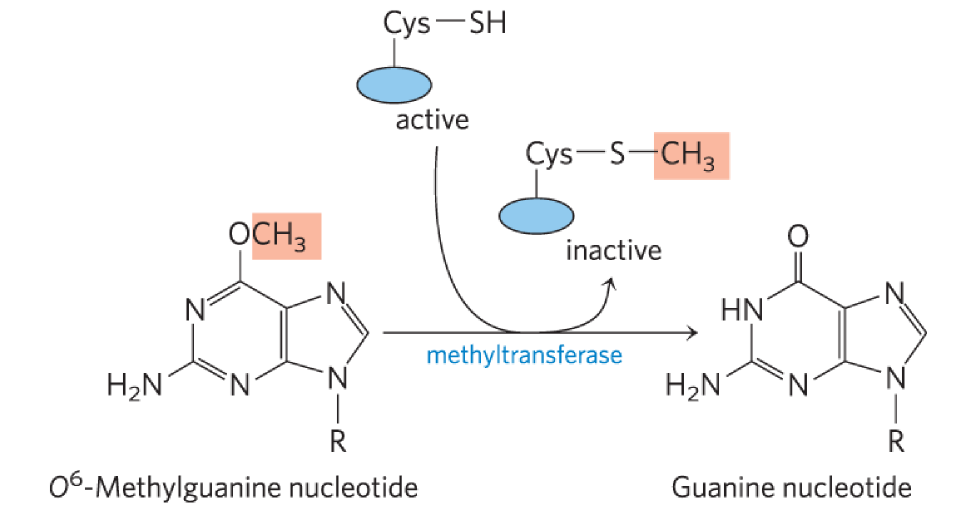
MGMT (O6-methylguanine-DNA methyltransferase)
functions in direct repair → fixes alkylation damage
catalyzes direct transfer of methyl group of O6-methylguanine to one of its own Cys residues
single methyl transfer event
Cys residue w/ thiol (SH) sidechain picks up methyl
permanently methylates protein → inactivated (suicide enzyme)
mechanism
binds to alkylated Guanine on minor groove side
binds w/ a-helical motifs, ie. Arginine
arginine = (+) → interacts w/ (-) DNA backbone → stabilizes reaction
another arginine allows damaged base to flip out
base goes into active site where methyl group can be removed → covalently attached to Cys
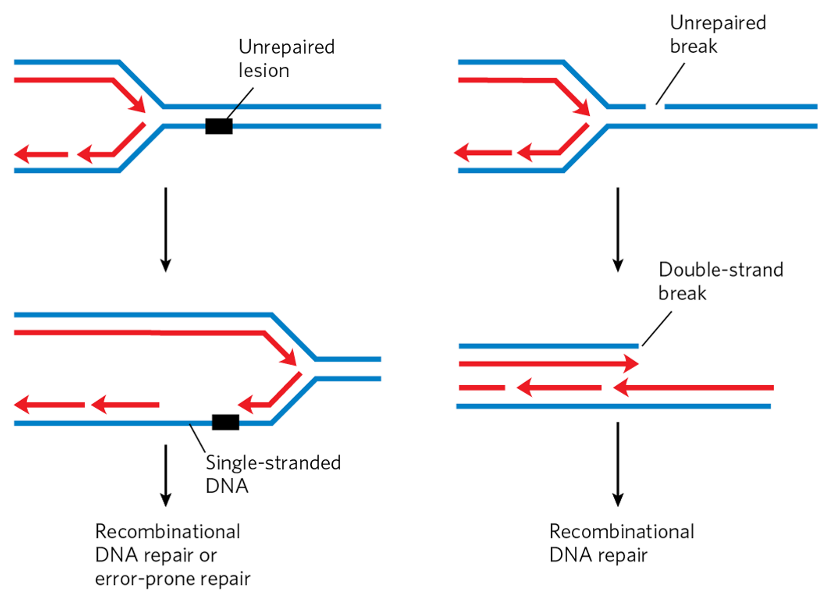
double strand breaks (DSBs)
caused by:
programmed DSBs
meiosis → recombination
V(D)J recombination
incomplete NER or BER repair + subsequent DNA replication
polymerase gets to end of linear chromosome but doesn’t continue until end
blunt-end break at unrepaired site
UV, IR or radiation damage
replication stalling
polymerase hits cross-link
can’t read + falls off
no Etna to bypass → polymerase rejoins across damage
single-strand region in newly synthesized strand
repair foci
large macro-molecular bodies that form when cells are damaged by IR/DSB
DNA repair proteins recruited and accumulate to facilitate repair
phosphorylation of H2A → yH2AX recruited to damage site
recruits protein 53BP1
binds to P53 → surveils genome
DSB detected = drives system towards apoptosis
no DSB detected = dissolves
PML-nuclear bodies dependent event → colocalize w/ persistent DNA damage foci
DSB repair pathways
HR
template-driven
conservative when sister chromosome used to repair break
NHEJ
join 2 broken strands w/ microhomology
can result in small deletions/insertions
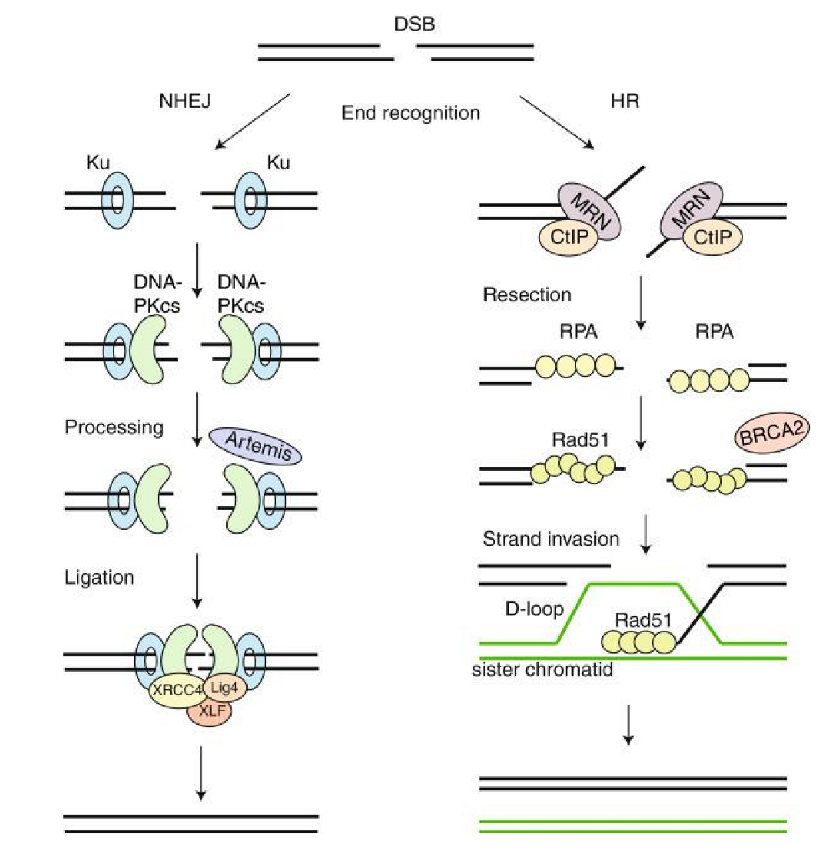
nonhomologous end joining (NHEJ)
favoured pathway in humans (except in S phase → HR would be favoured)
error prone → insertions + deletions if overhangs don’t align perfectly
mechanism
break recognition
Ku70-Ku80 complex binds DNA ends + recruits repair factors
help keep ends together to prevent strands from diffusing away
DNA-PKcs (kinase) bound to Artemis recruited
phosphorylates → kinase cascade
Artemis “processes” site
widens gap to allow more repair factors to bind
has nuclease + helicase activity
broken DNA ends synapsed by Artemis
on both sides of DSB, 1 strand of double helix lifted → flap
overhangs anneal via antiparallel complementary WC interaction
Artemis removes single-strand extensions/hairpins → end-processing
XLF + XRCC4 + ligase IV complex seals up phosphodiester backbone
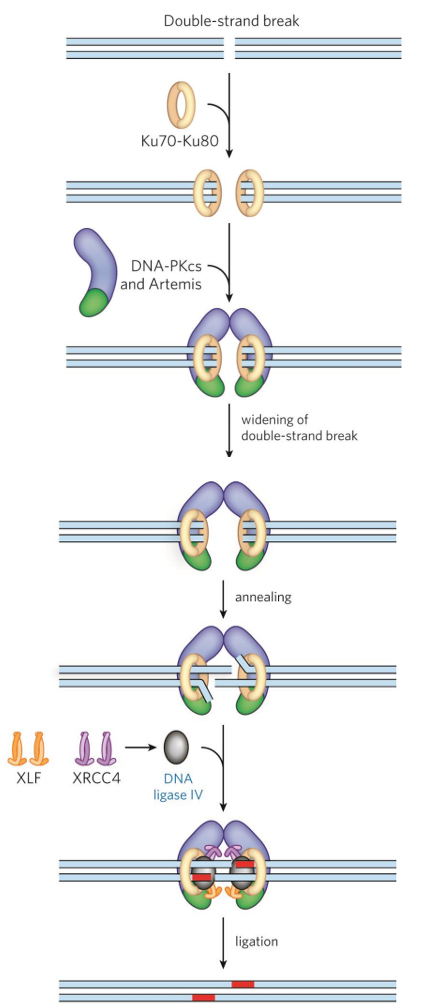
homologous recombination (HR) mechanism
nuclease resects DNA at 5’ end → large single-stranded DNA overhangs w/ 3’OH made
resection = commitment to repair pathway
RPA binds to ssDNA overhang
prevent strand from dipping into other DNA strands
BRCA2-mediated repair filament = traffic controller
assembles repair filament that can go into intact DNA strand → invasion filament
3’ overhang RPA displaced by RecA (E.coli) or RAD51 (humans) → becomes invasion filament
controlled by BRCA2
BRCA1/BARD1-mediated strand invasion
replaces BRCA2 + binds to RAD51 to form D-loop
3’ strand probes undamaged template for complementarity
if NT complementary → RAD51 peels away like zipper
criss-cross formed = Holliday junction
polymerase uses loop as template from free 3’OH as primer to fill in resected strands
second end capture
nuclease resolves Holliday junctions
ligase seals nicks
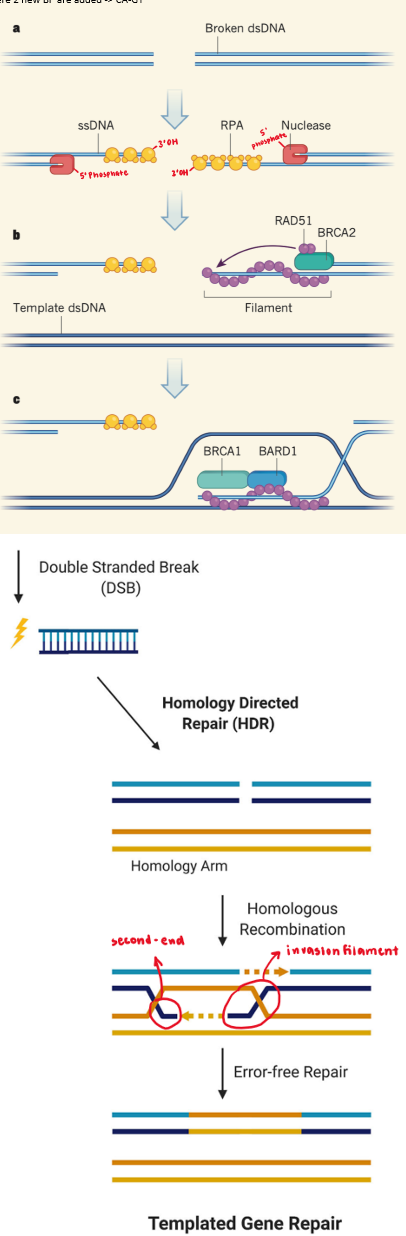
homologous recombination (HR)
high-fidelity repair pathway for DSBs in S or G2 phase
42% of human genome = line elements → repetitive
DNA broken in middle of line element → would go repair on any line element = ❌
translocation + fusion of 2 parts of chromosomes
humans spend lots of time in G1 = long growth phase
do not have identical copies in G1
could end up in G1 if repair pathway used
no insertions or deletions!
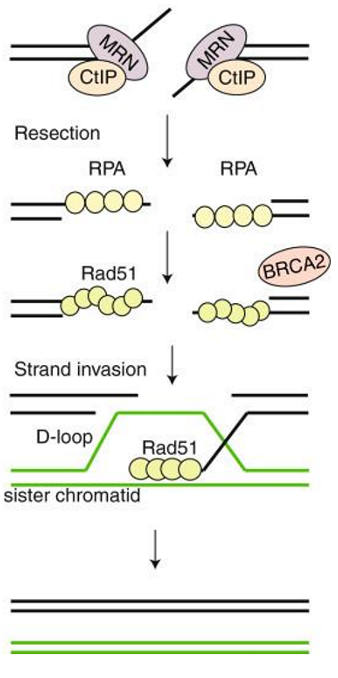
Holliday junction
structure formed during HR where 2 DNA strands exchange segments
resolved by nucleases
nuclease makes single-strand nicks in backbone
why HR can be used to swap around genes during meiosis
nicks sealed up by ligase
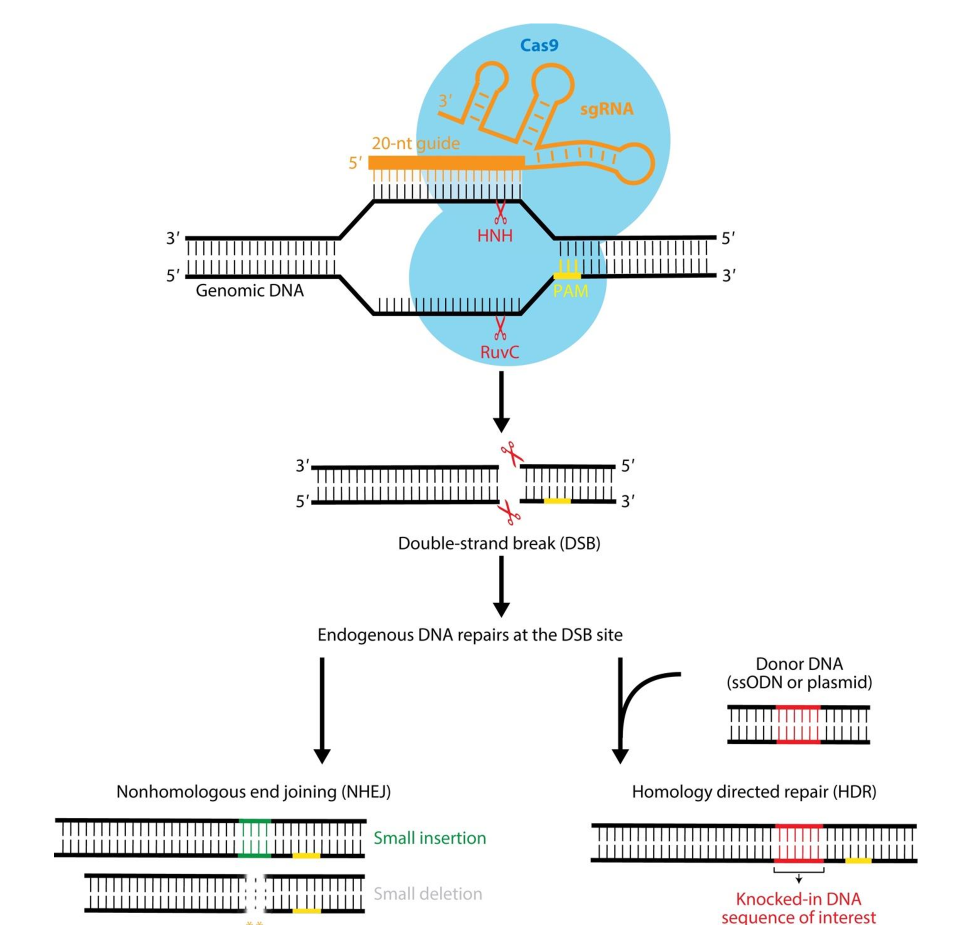
CRISPR-Cas9 mechanism
no RNA (CRISPR) = Cas9 protein in Apo (inactive) state
PAM recognition region = disordered
Cas9 binds to CRISPR RNA → activated
PAM recognition region = ordered
order PAM recognition region looks for PAM on target sequence
PAM = 5’-XGG-3’ sequence
RNA molecule parts
orange = structural component → binds to protein + fuels reorganization of PAM recognition site
white = directs traffic
purple = engineered sequence of interest → drives specificity
complementary to NTs across from PAM on target strand
1st purple NT makes antiparallel WC interaction w/ target strand → nucleation site
continues to unwind → more WC interactions
R-loop made → RNA-DNA complex
after 15-20 NTs make WC interactions → large structural reorganization of HNH domain
HNH moves up + over 3 NTs from PAM → active site opens + can cleave target strand
RuvC domain in open conformation
lines up 3NTs away from PAM on non-target strand (w/ PAM)
blunt DSB generated 3 NTs away from PAM + complex falls apart
RNA dissociates from DNA
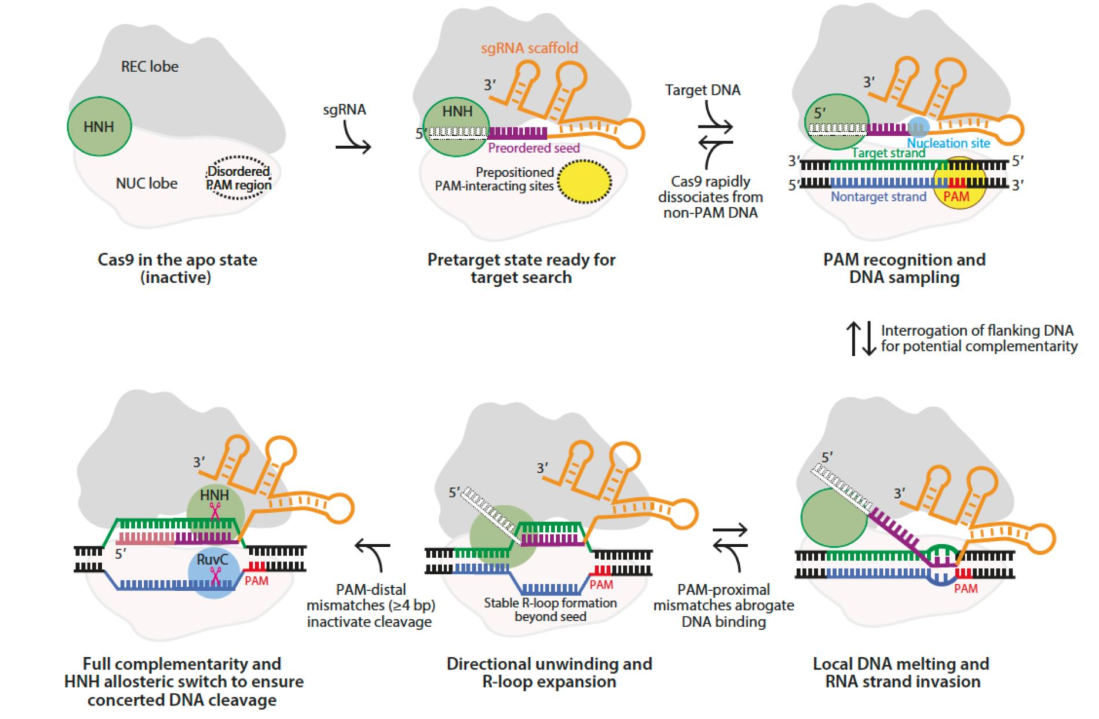
Cas9
main protein in CRISPR
3 domains
PAM recognition domain
disordered = can’t recognize PAM
ordered = can recognize PAM
binds to lots of PAMs but falls off if no complemntarity found w/ target sequence
HNH domain
nuclease cleaving template strand 3 NTs away from PAM
RuvC domain
nuclease cleaving non-template strand 3 NTs away from PAM
CRISPR-Cas9
genome editing technology to generate site-specific break
uses guide RNA + Cas9 protein to induce DSBs at specific locations in DNA
DSBs repaired by endogenous repair pathways
NHEJ
can result in small insertion/deletion → gene disruption
HR
template added → HR driven
suppress proteins involved in NHEJ + drive HR
can add DNA sequence of interest