BIOL 434 Exam 1
0.0(0)
0.0(0)
Card Sorting
1/134
Earn XP
Description and Tags
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
135 Terms
1
New cards
1\.List the four non-covalent interactions encountered in living systems
van der waals forces, hydrophobic interactions, hydrogen bonding, and dipole-dipole forces
2
New cards
1\.Explain the general characteristics of non-covalent interactions and the significance of such interactions in biological systems
solvents impact their strength, they have a cumulative effect that is highest in the native state, they decrease the free energy of the system while releasing binding energy
3
New cards
1\.Describe the origin of hydrogen bonds and give examples of molecules that form hydrogen bonds
N,O,S are attracted to H through partial charge differences
4
New cards
1\.Recognize the characteristics of hydrogen bonds and the importance of hydrogen bonds
water drives the movement of electrons across the membrane and interacts with amino acids through H bonding to drive molecular action- h bonds are stronger w/o water
they also form in between hydroxyl, carbonyl groups, and form between nucleotides
when the hydrogen acceptor is in a straight line with the H, the bond is stronger
they also form in between hydroxyl, carbonyl groups, and form between nucleotides
when the hydrogen acceptor is in a straight line with the H, the bond is stronger
5
New cards
1\.Explain what ionic interactions are and how they form
they are an attraction or repulsion of pos/neg charges, us. btwn Ca2+, phospho, carboxyl, amino groups
water stabilizes them by creating h-bonds around them which decreases the hydrostatic attraction around them
water stabilizes them by creating h-bonds around them which decreases the hydrostatic attraction around them
6
New cards
1\.Explain what is meant by the hydrophobic effect, how hydrophobic interactions occur, and their importance
includes the exclusion of polar bonds from a non-polar solution as non-p regions cluster to decrease the SA that interacts with the aq sol’n
water forms a highly ordered cage around to decrease the number of h bonds while the bulk phase around is less ordered
water forms a highly ordered cage around to decrease the number of h bonds while the bulk phase around is less ordered
7
New cards
1\.Define amphipathic molecule and explain what happens when such molecules are placed into water
partially hydrophobic/philic. will curl into a micelle to increase entrophy, philic is exposed and phobic is in the center
8
New cards
1\.Explain what Van der Waals forces are
all molecules do this as attractants and repulsions due to internal charges
9
New cards
Describe what weak acids and bases are
do not dissolve completely in sol’n
10
New cards
Explain what is meant by a conjugate acid-base pair and give examples of such pairs
acid donates a H making the conj base
acetic acid becomes acetate
carbonic acid becomes bicarbonate
acetic acid becomes acetate
carbonic acid becomes bicarbonate
11
New cards
Give examples of monoprotic, diprotic, and triprotic weak acids
mono:
acetic acid, pyruvic acid, lactic acid
di:
carbonic acid, succinic acid
tri:
phosphoric acid, citric, glutamic
acetic acid, pyruvic acid, lactic acid
di:
carbonic acid, succinic acid
tri:
phosphoric acid, citric, glutamic
12
New cards
Give the equation for the dissociation (ionization) constant, Ka
Ka= \[A-\]\[H+\]/\[HA\]
13
New cards
Describe what Ka is a measure of and how Ka is used to determine pKa
Ka tells how easily an acid loses a proton, where a higher Ka indicates a stronger acid
pH= pKa + log (\[A-\]/\[HA\])
pH= pKa + log (\[A-\]/\[HA\])
14
New cards
Explain what the pKa value of a weak acid indicates
a lower pKa indicates a stronger acid
when pKa = pH, half of the acid has dissolved in sol’n so it is half acid half base
when pKa = pH, half of the acid has dissolved in sol’n so it is half acid half base
15
New cards
Predict the predominant molecular species present in a solution containing a weak acid
at: pKa1- the carboxyl group is de protonated,
pKa2- amino group is deprotonated, pKa3- R group is ionized
as the pH increases, acids lose more protons
pKa2- amino group is deprotonated, pKa3- R group is ionized
as the pH increases, acids lose more protons
16
New cards
Describe what titration is and the information that a titration curve provides us
\
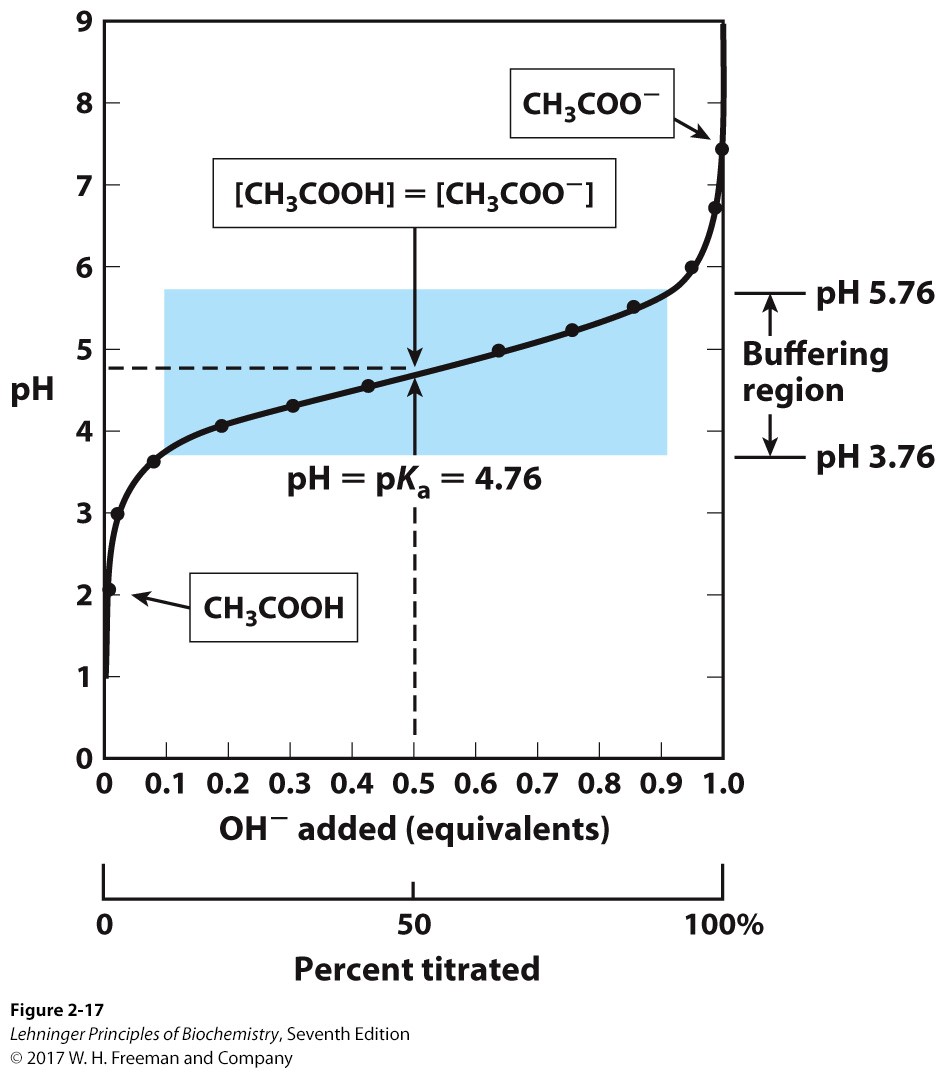
17
New cards
Explain what a buffer is
it is a sol’n made of an acid and its conjugate base that has a capacity of +/- 1 of the pKa
18
New cards
explain the importance and examples of buffers in biological systems, and consequences of malfunctions in buffering
phosphate and bicarbonate buffers are found in humans
carbonate becomes bicarbonate where carbonic anhydrase allows for Co2 to form which diffuses from capillaries to the lungs as CO2
when you exhale you release CO2 from the lungs which lowers the amount in the capillaries so equilibrium shifts to make more CO2 and hyperventilation occurs so kidneys reabsorb more CO2 into the blood
too much acid= acidosis, occurs from too much met byproduct, and too little= alkalosis from hyperventilation due to HCl leaking from the stomach or respiratory blockage
carbonate becomes bicarbonate where carbonic anhydrase allows for Co2 to form which diffuses from capillaries to the lungs as CO2
when you exhale you release CO2 from the lungs which lowers the amount in the capillaries so equilibrium shifts to make more CO2 and hyperventilation occurs so kidneys reabsorb more CO2 into the blood
too much acid= acidosis, occurs from too much met byproduct, and too little= alkalosis from hyperventilation due to HCl leaking from the stomach or respiratory blockage
19
New cards
Describe how buffers work
conj acid and base each donate or accept new protons to avoid drastic pH changes
20
New cards
Recognize the mathematical relationships between pH, pKa, and buffer concentration defined by the Henderson-Hasselbalch equation
pH=pKa + log(\[A-\]/\[HA\])
21
New cards
Solve various problems to determine pKa, pH, molar ratios, concentrations of compounds, and volumes to mix using the Henderson-Hasselbalch relationship
see hw sheet
22
New cards
Recognize and describe the structure of amino acids (AA) and recognize the different ways AAs are depicted
\:)
23
New cards
2\. Describe AA stereochemistry, the types of stereoisomers, and how D and L forms of AAs are determined
can be in L or D form (compared to Ala) amino group is on the left (left is more common) L and D are enantiomers
24
New cards
3\. Classify each of the 20 AAs commonly found in proteins into categories based on their R-groups
non-polar non-aromatic:
gly, ala, val, met, leu, pro, ile
(usually inside of the protein bc hydrophobic, gly is flexible)
\
aromatic:
trp, phe, try
\
polar uncharged:
ser, thr, cys, asn, gln
(csy makes disulfide bonds to stabilize 2o structure can form H bonds)
\
pos charge:
lys, arg, his
(not all pos at pH 7, arg can become guanidium, his has an imidazole ring, ionic interactions)
\
neg charge:
glu, asp
(ionic interactions and carboxyl makes it neg)
gly, ala, val, met, leu, pro, ile
(usually inside of the protein bc hydrophobic, gly is flexible)
\
aromatic:
trp, phe, try
\
polar uncharged:
ser, thr, cys, asn, gln
(csy makes disulfide bonds to stabilize 2o structure can form H bonds)
\
pos charge:
lys, arg, his
(not all pos at pH 7, arg can become guanidium, his has an imidazole ring, ionic interactions)
\
neg charge:
glu, asp
(ionic interactions and carboxyl makes it neg)
25
New cards
4\. Recognize and draw each of the 20 AAs commonly found in proteins as well as the AAs citrulline and ornithine
purr
26
New cards
5\. Give the three letter and the single letter abbreviation for each of the 20 AAs commonly found in proteins
purr
27
New cards
6\. Recognize that many more than 20 amino acids exist and some common AA residues in protein are modified post synthesis although some are introduced during protein synthesis
purr
28
New cards
Recognize selected modified AAs found in proteins and list in which proteins specific uncommon AAs are found
4-hydroxyproline and 5-hydroxylysine = collagen
6-N-methyllysine = myosin
gamma-carboxyglutamate = prothrombin
desmosine = elastin
selenocysteine = mod during synth from a UGA stop codon mod with own tRNA
6-N-methyllysine = myosin
gamma-carboxyglutamate = prothrombin
desmosine = elastin
selenocysteine = mod during synth from a UGA stop codon mod with own tRNA
29
New cards
8\. Recognize selected AAs that undergo reversible modification
phosphoserine, photyrosine, phosphothreonine
omega-n-methylarginine, 6-n-acetyllysine, glutamate y-methyl ester, adenylytyrosone
omega-n-methylarginine, 6-n-acetyllysine, glutamate y-methyl ester, adenylytyrosone
30
New cards
9. List AAs that are not found in protein but nonetheless have important biological functions
ornithine and citrulline are synthetic part of the urea cycle
thyroxine is a polytyrosine:
iodine + tyrosine with enzyme thyroperoxidase makes thyroglobin, monoidenotyrosenase and di, add I to T’s to make T3 and T4
glutamate is decarboxylated to GABA
his is decarboxylated to histamine
thyroxine is a polytyrosine:
iodine + tyrosine with enzyme thyroperoxidase makes thyroglobin, monoidenotyrosenase and di, add I to T’s to make T3 and T4
glutamate is decarboxylated to GABA
his is decarboxylated to histamine
31
New cards
10\. Explain why AAs have ionic properties and utilize the terms amphoteric and zwitterion to describe AAs
they have a pos and neg end that can change as the pH of the sol’n changes as AA are amphoteric. When at the pI point, the AA is a zwitterion or pos and neg at the same time but electrically neutral
32
New cards
11\. Describe the general ionization patterns of AAs, sketch the titration curve for various AAs, and identify/label points on the curve such as where all species are fully protonated or deprotonated, the isoelectric point (pI), where the AA is a zwitterion, inflection points
screaming check the ppt
33
New cards
12\. Define peptides, draw a simple peptide when given the name of the peptide, and name the peptide when it is already drawn out
screaming
34
New cards
Recognize that peptides can ionize and predict the overall charge of a short peptide at a given pH
beep
35
New cards
14\. List peptides of physiological interest and what role those peptides play
lamosine= alanly serine dipeptide
enkephalins= pentapeptides that bind like opiates to receptors
oxytocin= nonapeptide that forms a disulfide bond and cyclic structure
antibiotics w/ 2 chains, gramicidin s and tyrocidine A are a cyclic mixture of D and L
enkephalins= pentapeptides that bind like opiates to receptors
oxytocin= nonapeptide that forms a disulfide bond and cyclic structure
antibiotics w/ 2 chains, gramicidin s and tyrocidine A are a cyclic mixture of D and L
36
New cards
List the four levels of protein structure
primary secondary tertiary quaternary
37
New cards
2\. Explain what is meant by conformation and native state of a protein
conform is rotation around a cov/ peptide bond and the native state maximizes the non-covalent interactions and lowers the free E
38
New cards
3\. Explain why only one or a very few possible conformations exist for protein under typical biological conditions
only a few can be thermodynamically the most stable due to the decrease in free E
39
New cards
4\. Describe the ways that conformation is stabilized
non-cov interactions, disulfide bonds (intracellular environment isn’t good for making DSB)
max weak interactions is the most stable and lowers free E
hydrophobic interactions do the most to stabilize proteins bc they result in the most increase in entropy of the universe, polar charged aa have to be balanced by aa of opposite charge, water doesn’t interfere with internal environment
max weak interactions is the most stable and lowers free E
hydrophobic interactions do the most to stabilize proteins bc they result in the most increase in entropy of the universe, polar charged aa have to be balanced by aa of opposite charge, water doesn’t interfere with internal environment
40
New cards
5\. Define what is meant by 1°structure, why knowing 1°structure is useful, and how 1°structure is a determinant of conformation
is AA sequence, used to tell mech of action of a protein for disease pathology, evolution, ect
41
New cards
6\. Describe what is meant by polymorphism
2 similar forms of same AA sequence (like HbA and HbS of hemoglobin) dift tissues might have different forms and shouldn’t dicrease function
42
New cards
7\. Describe the peptide bond backbone and the various bonds and bond angles involved
peptide bond is planar and backbone is planar in trans configuration bc of pos and neg charges
R groups would clash if cis (pro has some cis)
are specific bond angles for dift types
N-alpha and Calpha- C can rotate but C-N don’t rotate (peptide bond)
impact secondary structure
R groups would clash if cis (pro has some cis)
are specific bond angles for dift types
N-alpha and Calpha- C can rotate but C-N don’t rotate (peptide bond)
impact secondary structure
43
New cards
8\. Define what is meant by 2°structure
the local spacial arrangement for main chain atoms (not R group)
44
New cards
what are the three major types of 2°structure
a-helix, b-conformation/b-pleated sheet, b-turn
45
New cards
what are the specific characteristics of alpha-helix
r groups protrude outside of the helix
bp are 1.5A apart and 3.6 res to make 1 helix
are elastic
form between the H bonds of amide and the O of the carbonyl group
(L amino and D aa make up the helix and if added separately will disrupt structure)
are us right handed (amino end points right)
some amino acids are more likely to form a helix
(aa that decrease change in delta G are more likely to form a helix) bc charged R-groups might repel or be too big and disrupt structure (can’t have 2+ adjacent)
amino term is pos carboxyl is neg
bp are 1.5A apart and 3.6 res to make 1 helix
are elastic
form between the H bonds of amide and the O of the carbonyl group
(L amino and D aa make up the helix and if added separately will disrupt structure)
are us right handed (amino end points right)
some amino acids are more likely to form a helix
(aa that decrease change in delta G are more likely to form a helix) bc charged R-groups might repel or be too big and disrupt structure (can’t have 2+ adjacent)
amino term is pos carboxyl is neg
46
New cards
what are specific char of beta sheets
made of 2-12 strands (us 6aa per strand)
polypep bb extends into a zig zag
3\.5A btwn aa (larger gap than alpha h)
R groups are on opposite sides
2+ strands are linked via H bonds based on parallel or anti, arrow shows the carboxyl end
bond angles change depending
anti= each aa forms 1 H bond with adjacent aa
par= each aa forms 2 h bonds with adjacent aa
polypep bb extends into a zig zag
3\.5A btwn aa (larger gap than alpha h)
R groups are on opposite sides
2+ strands are linked via H bonds based on parallel or anti, arrow shows the carboxyl end
bond angles change depending
anti= each aa forms 1 H bond with adjacent aa
par= each aa forms 2 h bonds with adjacent aa
47
New cards
what are specific char of beta turns
polypeptide reverses direction (180degree turn) anti-prolyl connectors
4 amino acids make up each one
type 1: make up the second residue (pro) and is not flexible
type 2: third res (gly) is flexible
form middle H bonds to hold turn in place
proline cis-isomer forms the turn (bc trans is straight)
4 amino acids make up each one
type 1: make up the second residue (pro) and is not flexible
type 2: third res (gly) is flexible
form middle H bonds to hold turn in place
proline cis-isomer forms the turn (bc trans is straight)
48
New cards
0\. Predict the 2°structure of a short sequence of amino acids
for alpha helix: amino term is pos carboxyl is neg so r-groups will oppose those charges, pro will not form alpha, gly is V flexible so will
for beta sheets: if multisheets are stacked, gly + ala are found at the surface bc they are small while large aromatic branched aa are us in the middle, pro on edges
for beta sheets: if multisheets are stacked, gly + ala are found at the surface bc they are small while large aromatic branched aa are us in the middle, pro on edges
49
New cards
Define / Describe what is meant by 3°structure and how a protein would be held together
conformation of the backbone and the r-groups
alpha helix crosslinked by DS bonds, beta conformation, collagen triple helix
alpha helix crosslinked by DS bonds, beta conformation, collagen triple helix
50
New cards
2\. Describe what is meant by prosthetic group, conjugated / holoprotein, metalloprotein, glycoprotein, lipoprotein
non-peptide structure required for tert structure to form
conjugated/ holoprotein have something intertwined with the protein, metallo= metal group, glyco= sugar, lipo= fat
conjugated/ holoprotein have something intertwined with the protein, metallo= metal group, glyco= sugar, lipo= fat
51
New cards
3. Describe the two major groups of proteins including their characteristics and their typical functions
globular: have lots of secondary structure types like alpha and beta, us enzyme, receptor, transport protein (like hemoglobin)
fibrous: hydrophobic, noncov, long, sheets, shape support external protection function (like keratin)
fibrous: hydrophobic, noncov, long, sheets, shape support external protection function (like keratin)
52
New cards
4\. Describe the structural patterns of globular proteins including: a. The definition of motif and different types of motifs
b. The basic rules of folding
c. The four different structural classifications based on motif
b. The basic rules of folding
c. The four different structural classifications based on motif
motif= folding patterns (super secondary structure) w/ 2+ elements
beta-alpha-beta loop= can make an alpha beta barrel
beta barrel
TIM barrel= alpha outside beta strand inside
all beta= no cross-over connections seen, us. right hand,
twisted beta sheets are more stable
some are all alpha folding, some are mixed
beta-alpha-beta loop= can make an alpha beta barrel
beta barrel
TIM barrel= alpha outside beta strand inside
all beta= no cross-over connections seen, us. right hand,
twisted beta sheets are more stable
some are all alpha folding, some are mixed
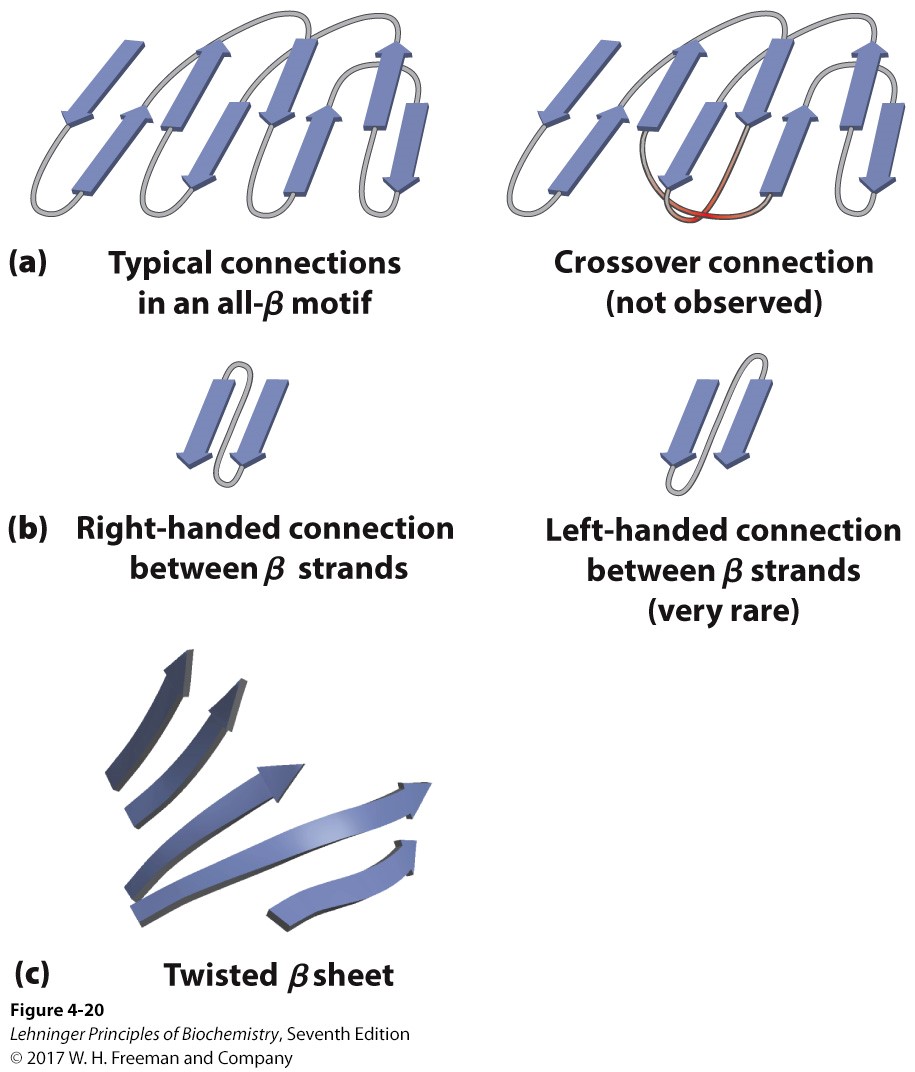
53
New cards
d. The difference between family and superfamily
family= high degree of similarity btwn primary and or tertiary structure and function
superfamilies= 2+ families with a high degree of similarity in motif and function, but less similarity of primary structure
superfamilies= 2+ families with a high degree of similarity in motif and function, but less similarity of primary structure
54
New cards
e. What is meant by domain
are parts that are independently stable in a protein
pep > than aa in size, can fold into 2+ domains w/ separate functions
more efficient for cat. 1 part may be for the AS while 1 is regulatory
1 subunit can regulate entire protein
pep > than aa in size, can fold into 2+ domains w/ separate functions
more efficient for cat. 1 part may be for the AS while 1 is regulatory
1 subunit can regulate entire protein
55
New cards
5\. Define / Describe what is meant by 4°structure, the various types of 4°structure, and the roles such proteins play in a cell
multi peptides with multi subunits
us. have complex multistep functions
can do cat. of things that are different but relate to the same function
us act unit by unit
us. have complex multistep functions
can do cat. of things that are different but relate to the same function
us act unit by unit
56
New cards
Explain what is meant by intrinsically disordered proteins, how their properties differ from structured proteins, and some of their recognized functions
intrinsically disordered proteins lack hydrophobic cores
not all proteins are entirely defined
have an increased density of charged AA
ranked with PONDR (predictor of naturally disordered regions) score
can be scavengers = bind to ions, small molecules, reservoirs, garbage dump, insulators, inter rxn networks
p27= main cell protein kinase to inhibit cell division (would find lower \[\] in tumor cells)
not all proteins are entirely defined
have an increased density of charged AA
ranked with PONDR (predictor of naturally disordered regions) score
can be scavengers = bind to ions, small molecules, reservoirs, garbage dump, insulators, inter rxn networks
p27= main cell protein kinase to inhibit cell division (would find lower \[\] in tumor cells)
57
New cards
Explain what is meant by proteostasis including pathways that contribute to proteostasis
proteostasis= the continual maint. of proteins in a cell
synthesis, folding, native state, unfolding, refolding, denaturation
synthesis, folding, native state, unfolding, refolding, denaturation
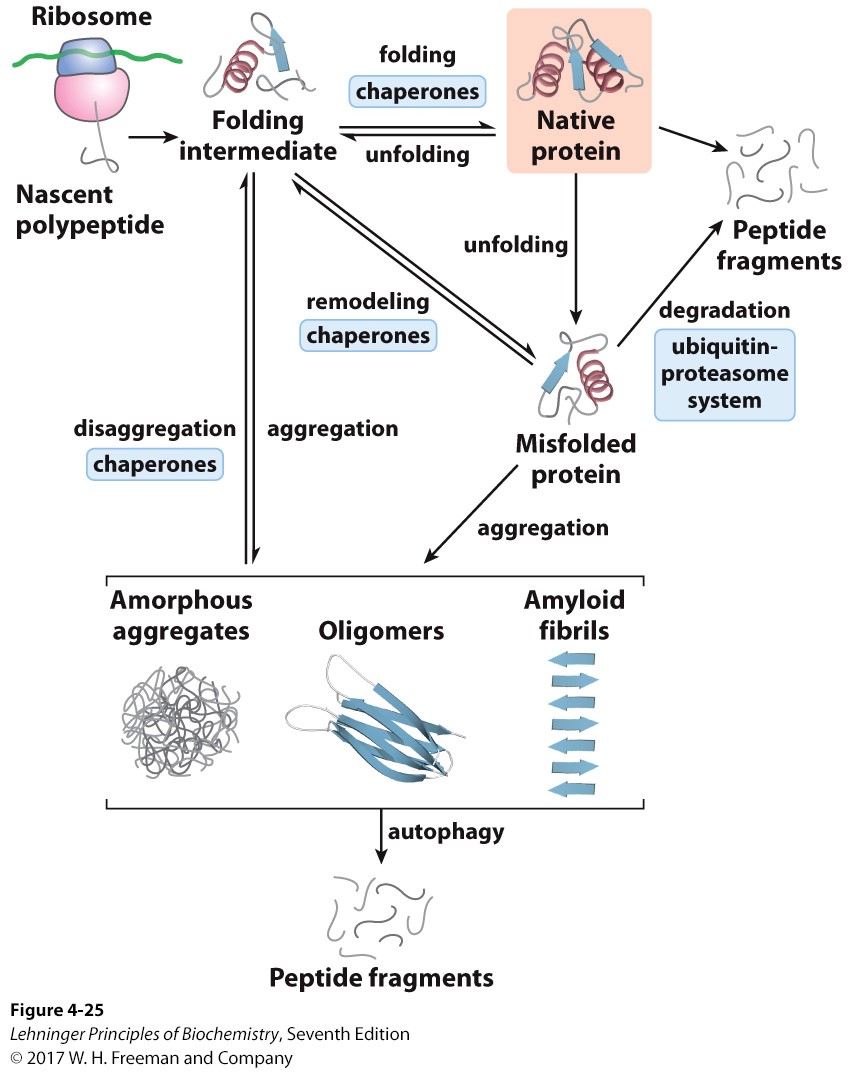
58
New cards
2\. Describe what occurs during denaturation of a protein and explain how heat, pH, organic solvents, and detergents result in denaturation of a protein
loss of 3D structure/ conformation decrease function in denaturing
temperature disrupts H-bonds
loss of H+ changes charge which causes denaturation
organic solvents (etOH, acetone)/detergents change hydrophobic interactions
temperature disrupts H-bonds
loss of H+ changes charge which causes denaturation
organic solvents (etOH, acetone)/detergents change hydrophobic interactions
59
New cards
3. Define renaturation
if DS bonds are broken into Cys res by urea/ mercaptoethanol they can be put in an environment that removes those toxins so the DS bonds can reform back to active/native state
60
New cards
4\. Describe how polypeptides / proteins are folded including hierarchical modeling
from primary structure, secondary forms next, alpha helix forms faster, beta sheets, weak interactions are the most important then ionic, finally domains are combined
61
New cards
what are 2 enzymes often needed for folding
PDI (protein disulfide isomerase) needed for folding sometimes
PPI (peptidyl proyl cis-trans isomerase- converts btwn cis and trans forms of proline)
PPI (peptidyl proyl cis-trans isomerase- converts btwn cis and trans forms of proline)
62
New cards
the role of thermodynamics in protein folding
therm is represented by funnels where the bottom shows the native lowest free E state, raised areas show stability of intermediates/ possible conformations, peaks show less stable intermediates but where present show more stability
63
New cards
what is the role of molecular chaperones
chap= react w/ partially or unfolded proteins to create a microenvironment for folding, also block newly formed proteins from aggregating, keeps proteins separate
64
New cards
what is an HSP
HSPs (heat shock proteins) prevent proteins from aggregating under high temperature
65
New cards
what do the examples of HSPs in e. coli do
in e. coli, HSP70 is open when ATP is bound and has low affinity, but closes when HSP40 binds and ATP leaves which then increases affinity to move hydrophobic regions of proteins together to prevent aggregation
66
New cards
5. Describe what a proteasome is,
large protein complexes that degrade unneeded or damaged proteins (found in most orgs)
67
New cards
the basic structure of the 26s eukaryotic proteasome,
top and bottom is 19s subunit that houses the docking site where polyubiq with protein attaches to the proteosome
alpha subunits next to it are where ATPases are and need ATP to work
beta in the middle is the active site where catalysis happens
alpha and beta subunits make up the 20s core
alpha subunits next to it are where ATPases are and need ATP to work
beta in the middle is the active site where catalysis happens
alpha and beta subunits make up the 20s core
68
New cards
how it works to break down a protein (proteolysis),
(4+ ubiquine) polyubiquinated proteins move the protein to the proteosome
enter the docking site, ATP enters the ATPases at the alpha subunit, protein moves to beta subunit
ubiquitin is removed then unfolding happens
long seq of ala/ gal inhibit unfolding
enter the docking site, ATP enters the ATPases at the alpha subunit, protein moves to beta subunit
ubiquitin is removed then unfolding happens
long seq of ala/ gal inhibit unfolding
69
New cards
what sites are needed in the beta subunit to cleave sites on a protein
sites for threonine dependent nucleophilic attacks to break proteins in certain locations
chymotrypsin like- cleaves peptide bonds on CA side of trp, try, phe (aromatic)
trypsin like- cleaves pep bond on CA side of lys and arg (except when pro is next!)
peptidyl glutnyl- hydrolyzing like- cleaves CA bond immediately after acidic or branch chain aa
chymotrypsin like- cleaves peptide bonds on CA side of trp, try, phe (aromatic)
trypsin like- cleaves pep bond on CA side of lys and arg (except when pro is next!)
peptidyl glutnyl- hydrolyzing like- cleaves CA bond immediately after acidic or branch chain aa
70
New cards
6\. Explain how a protein is targeted (or not) for degradation
Degron (amino end terminal) signals for faster or slower degradation, PEST (pro glut ser threo) signals, TF Grn4P reduces 1/2 life too
intrinsically disordered proteins don’t need a signal but Cyclin B does
intrinsically disordered proteins don’t need a signal but Cyclin B does
71
New cards
name the three enzymes and their functions in the ubiquitination process
E1= ubiquitin activating enzyme
E2= ubiquitin conjugating enzyme
E3= ubiquitin ligase
carboxyl group reacts with SH group on E1 and ATP to give off pyrophosphate
E2 transfers itself where E1 was
reacts with E3 and amino end of Lys of the target protein so that now enzymes are gone but target protein attaches (does this 4 times thru ubiquitin elongation)
ligase attaches to epsilon aa
E2= ubiquitin conjugating enzyme
E3= ubiquitin ligase
carboxyl group reacts with SH group on E1 and ATP to give off pyrophosphate
E2 transfers itself where E1 was
reacts with E3 and amino end of Lys of the target protein so that now enzymes are gone but target protein attaches (does this 4 times thru ubiquitin elongation)
ligase attaches to epsilon aa
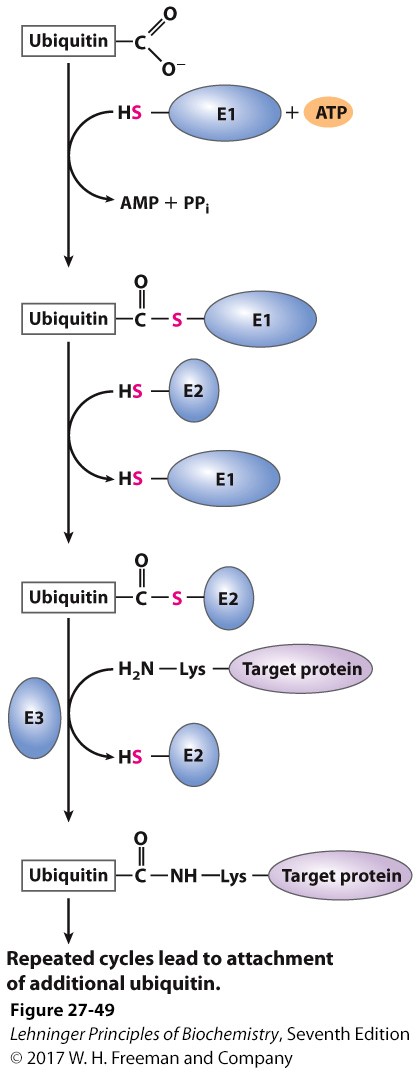
72
New cards
7\. Describe what amyloidoses are and give specific examples that are known or suspected
E3 malfunction so can’t attach target protein to ubiq
(HPV has too much ubiq so destroys p53)
disease from improper folding/ misfolding
insoluble fibers (amyloid fibers/ plaques) increase the number of ordered structure like beta sheets so abnormally folded proteins recruit correctly folded proteins into the wrong form
type 2 diabetes can be caused by amylin when secreted by the pancreas it has a synergistic affect with insulin and forms fibers so beta cells apoptosis and insulin decreases
alzheimer’s has increased levels of beta-amyloid fibers
amyloid pre-cursor in neurons is the protein APP which which can be proteolyzed which causes prion diseases which result from an increase in beta sheets and alpha helix
parkinsons is a maybe
(HPV has too much ubiq so destroys p53)
disease from improper folding/ misfolding
insoluble fibers (amyloid fibers/ plaques) increase the number of ordered structure like beta sheets so abnormally folded proteins recruit correctly folded proteins into the wrong form
type 2 diabetes can be caused by amylin when secreted by the pancreas it has a synergistic affect with insulin and forms fibers so beta cells apoptosis and insulin decreases
alzheimer’s has increased levels of beta-amyloid fibers
amyloid pre-cursor in neurons is the protein APP which which can be proteolyzed which causes prion diseases which result from an increase in beta sheets and alpha helix
parkinsons is a maybe
73
New cards
Recognize and describe different methods of protein isolation/separation and characterization including: a. Salting out
1st- make crude extract (cell lysis)
2- homogenize material
3- use differential centrifugation to make subcellular fractions
4- fractionations- separate proteins in groups by density
salting out: ppt out proteins in a sol’n of (NH4)2SO4 and centrifuge to get a pellet
2- homogenize material
3- use differential centrifugation to make subcellular fractions
4- fractionations- separate proteins in groups by density
salting out: ppt out proteins in a sol’n of (NH4)2SO4 and centrifuge to get a pellet
74
New cards
Dialysis
separate protein from small solutes by putting them in a bag of a semi-permeable membrane that will let the small solutes diffuse out leaving the protein behind
75
New cards
c. Column chromatography
shows the protein’s charge, size, and capacity to bind to other molecules
protein is in reservoir where it is pumped in a buffer to columns where higher affinity proteins stay at the top while lower affinity sub move faster and have a lower retention time
when moving it is in the mobile phase and when attached to the porus matrix it is in the stationary phase
a detector notes to a recorder and a fraction collecter collects to liquid
doing all types will increase the activity (total units of an enzyme in sol’n/amount of space, high activity= more pure)
protein is in reservoir where it is pumped in a buffer to columns where higher affinity proteins stay at the top while lower affinity sub move faster and have a lower retention time
when moving it is in the mobile phase and when attached to the porus matrix it is in the stationary phase
a detector notes to a recorder and a fraction collecter collects to liquid
doing all types will increase the activity (total units of an enzyme in sol’n/amount of space, high activity= more pure)
76
New cards
ion exchange column chromatography
shows differences in magnitudes of charges
keep buffer at a certain pH (can change pH to release if know pI)
allow protein to move down
beads (resin is neg) are charged and opposite charges move most slowly while same move more quickly
keep buffer at a certain pH (can change pH to release if know pI)
allow protein to move down
beads (resin is neg) are charged and opposite charges move most slowly while same move more quickly
77
New cards
size-exclusion column chrom
porus beads allow for small proteins to go through tunnels in them so they move more slowly down the column than larger beads
78
New cards
affinity column chrom
ligand is attached to beads and protein is allowed to go down and speed is determined by affinity for ligand
79
New cards
high performance liquid chromatography (HPLC)
uses pressure pumps to move protein down at a specific flow rate
80
New cards
d. Electrophoresis
used to separate and analyze but not purify bc enzyme will no longer be in native state when done
uses charge and size, migration speed is directly proportional to strength of the net charge of molecules
polyacrimide gel electrophoresis (PAGE) is used with detergent
simple PAGE keeps it in the native state but SDS PAGE disrupts the non-cov bonds (1:1 SDS: protein molecule ration)
makes a uniform rod shape and denatures so original charge doesn’t matter
so each protein will have a similar charge:mass ratio so bigger molecules will be more negative and move faster
more purified protein will have a more defined band
can be used to make a standard curve
uses charge and size, migration speed is directly proportional to strength of the net charge of molecules
polyacrimide gel electrophoresis (PAGE) is used with detergent
simple PAGE keeps it in the native state but SDS PAGE disrupts the non-cov bonds (1:1 SDS: protein molecule ration)
makes a uniform rod shape and denatures so original charge doesn’t matter
so each protein will have a similar charge:mass ratio so bigger molecules will be more negative and move faster
more purified protein will have a more defined band
can be used to make a standard curve
81
New cards
e. Isoelectric focusing
separates protein by pI values. puts protein in a pH and electrical field gradient and will stop moving when reaches its pI point bc electrically neutral
82
New cards
f. Two dimensional electrophoresis techniques
do isoelectric focusing and put on a strip on gel and run it on SDS PAGE thru electrophoresis to sep by pI and size each dot seen represents a protein
83
New cards
2\. Explain what is meant by activity and specific activity
84
New cards
3\. Describe how amino acid sequence/composition and structure can be determined
use purified sequence
can tag amino end using FDNB which cleaves at the amino end of Lys to breakdown larger peptides
use Cyanogen Br to cleave the C end of Met
use dift proteases to bd protein
put the sequences together
\
edmund degredation sequences aa are cleaved and ID’d one by one
mass spec finds a mass: charge ration for each fragment
can tag amino end using FDNB which cleaves at the amino end of Lys to breakdown larger peptides
use Cyanogen Br to cleave the C end of Met
use dift proteases to bd protein
put the sequences together
\
edmund degredation sequences aa are cleaved and ID’d one by one
mass spec finds a mass: charge ration for each fragment
85
New cards
which amino acids do the following reagents cleave on the C terminus
trypsin
submaxillary protease
chymotrypsin
S. areus
cyanogen bromide
trypsin
submaxillary protease
chymotrypsin
S. areus
cyanogen bromide
Lys Arg
Arg
aromatics
Asp Glu
Met
Arg
aromatics
Asp Glu
Met
86
New cards
which amino acids do the following reagents cleave on the N terminus
Asp-N-Protease
Pepsin
Asp-N-Protease
Pepsin
Asp Glu
Leu + aromatics
Leu + aromatics
87
New cards
how to measure activity
monitor unknown rxn and compare to known rxn with a standard curve to see how active it is
88
New cards
find protein \[\]
use spec and Lowry argent for color
Coomassie blue makes it all blue dye loses protons when bound, so can make standard curve to see \[ \]
Coomassie blue makes it all blue dye loses protons when bound, so can make standard curve to see \[ \]
89
New cards
5\. Solve problems utilizing the various methods
slay
90
New cards
Describe the common aspects of non-enzymatic functions of proteins particularly interactions involving changes in conformation caused by ligand binding to a protein.
binding to a ligand us. is reversible due to conformational changes
protein AS are specific in size, charge, and shape
binding sites become more complimentary to protein after a conform change causing an induced fit so the ligand and protein bind more tightly
if there are multi sub for one protein, they will work in a regulatory process where the conformational change of the first allows for the next to work
protein AS are specific in size, charge, and shape
binding sites become more complimentary to protein after a conform change causing an induced fit so the ligand and protein bind more tightly
if there are multi sub for one protein, they will work in a regulatory process where the conformational change of the first allows for the next to work
91
New cards
describe the structure of myoglobin (which is monomeric)
has 8 alpha helix subunits and one heme group
it has a heterocyclic ring derived from porphyrin with 4 pyrole groups
protein portions prevent permanent binding but when removed, binding is permanent
heme group interacts with 4 N ligands in they pyrole groups that are found on the same plane, the 5th ligand interacts with the His93 imidazole side chain which slightly displaces the Fe from its plane, the 6th ligand will bind to O2
it has a heterocyclic ring derived from porphyrin with 4 pyrole groups
protein portions prevent permanent binding but when removed, binding is permanent
heme group interacts with 4 N ligands in they pyrole groups that are found on the same plane, the 5th ligand interacts with the His93 imidazole side chain which slightly displaces the Fe from its plane, the 6th ligand will bind to O2
92
New cards
how does myoglobin work
when O2 binds to the 6th ligand it pulls back from the plane when not bound
Fe2+ can be oxidized to Fe3+ or metmyoglobin
metmyoglobin can no longer bind to O2
CO and H2S bind to myoglobin better than O2
its O2 sat. curve is a regular MM curve because it is monomeric and thus does not bind O2 cooperatively
is not as sensitive to changes in O2 and better for O2 storage bc of high O2 affinity
Fe2+ can be oxidized to Fe3+ or metmyoglobin
metmyoglobin can no longer bind to O2
CO and H2S bind to myoglobin better than O2
its O2 sat. curve is a regular MM curve because it is monomeric and thus does not bind O2 cooperatively
is not as sensitive to changes in O2 and better for O2 storage bc of high O2 affinity
93
New cards
describe the structure of hemoglobin (which is tetrameric)
subunits alpha 1 and beta 1 interact well and alpha 2 and beta 2 interact well but alpha/beta 1 and alpha/beta 2 don’t
has 2 conformations
T state (unbound) R state (bound)
T= his HC3 part is found on the outside
R= his HC3 found in the center
R has a higher affinity for O2 than T
has 2 conformations
T state (unbound) R state (bound)
T= his HC3 part is found on the outside
R= his HC3 found in the center
R has a higher affinity for O2 than T
94
New cards
how does hemoglobin work
in lungs: in T state where binds to 1 O2, and conform change allows for rest of O2 to bind
enters the R state
goes to the body tissues: where it looses 1 O2 and then a conformational change allows for all others to leave
enters the T state
T state is more stable than R state where subunits slide and rotate more easily for conformational changes
its O2 sat. curve is a sigmoidal MM curve because it is tetrameric and can bind O2 cooperatively
is more sensitive to O2 changes and better for O2 transport bc of lower O2 affinity
enters the R state
goes to the body tissues: where it looses 1 O2 and then a conformational change allows for all others to leave
enters the T state
T state is more stable than R state where subunits slide and rotate more easily for conformational changes
its O2 sat. curve is a sigmoidal MM curve because it is tetrameric and can bind O2 cooperatively
is more sensitive to O2 changes and better for O2 transport bc of lower O2 affinity
95
New cards
Explain how Kd is derived and the information that Kd provides
if Y= total number of binding sites occupied/ total binding sites aka ( \[PL\]/(\[PL\] +\[P\]))
Ka is the association constant and Ka \[L\]\[P\] can be sub for \[PL\]
at Y=.5 half of the binding sites are occupied and is known as 1/Ka or Kd (M) Kd= kd/ka
Y= \[L\]/(\[L\] + Kd)
**Kd= [P]*[L]/[PL]**
therefore when \[L\] = Kd half of the ligand binding sites are occupied
when \[L\] is less than Kd, less protein is bound to ligand
lower Kd= higher affinity of ligand for protein
the more tightly a protein binds to a ligand, the lower concentration of ligand is needed for half of the binding sites to be occupied which means a lower Kd
Ka is the association constant and Ka \[L\]\[P\] can be sub for \[PL\]
at Y=.5 half of the binding sites are occupied and is known as 1/Ka or Kd (M) Kd= kd/ka
Y= \[L\]/(\[L\] + Kd)
**Kd= [P]*[L]/[PL]**
therefore when \[L\] = Kd half of the ligand binding sites are occupied
when \[L\] is less than Kd, less protein is bound to ligand
lower Kd= higher affinity of ligand for protein
the more tightly a protein binds to a ligand, the lower concentration of ligand is needed for half of the binding sites to be occupied which means a lower Kd
96
New cards
Predict which molecule has a higher affinity for a protein when the Kd for various ligands is known
lower Kd means higher affinity
97
New cards
Recognize the structure of a heme including the different components
iron in the center w/ 4 pyrrole groups around it
Fe is planar w/ pyrrole groups when no O2 is bound and impacts the distance btwn the distal His
Fe is planar w/ pyrrole groups when no O2 is bound and impacts the distance btwn the distal His
98
New cards
Distinguish between different models of cooperativity
cooperative binding is seen when a sigmoidal curve is made and means that the complex can be more sensitive to small changes in O2
MWC says that all substrates at any given time are in the same conformation so an equal chance for ligand to bind to any (all or none)
cooperative binding in sequential method says that a conformational change in one subunit makes it more likely to be bound which then impacts the binding of the next subunit in a more sequential manner
MWC says that all substrates at any given time are in the same conformation so an equal chance for ligand to bind to any (all or none)
cooperative binding in sequential method says that a conformational change in one subunit makes it more likely to be bound which then impacts the binding of the next subunit in a more sequential manner
99
New cards
Describe the role BPG plays in O2 binding
hemoglobin has binding sites for CO2 and H+
CO2 needs to be converted to bicarb via carbonic anhydrase or it would bubble out this means that the pH will decrease in this process
when pH in tissues decreases, more O2 can be released so the affinity for O2 will increase in the lungs
BPG binds to the T state to stabilize it so that the affinity for O2 is lower so that at high altitudes O2 can be released (a person from higher altitudes will have more BPG)
CO2 needs to be converted to bicarb via carbonic anhydrase or it would bubble out this means that the pH will decrease in this process
when pH in tissues decreases, more O2 can be released so the affinity for O2 will increase in the lungs
BPG binds to the T state to stabilize it so that the affinity for O2 is lower so that at high altitudes O2 can be released (a person from higher altitudes will have more BPG)
100
New cards
Identify heme (not including myosin and hemoglobin) based O2 binding protein as well as non-heme O2 binding protein, indicate where they are found, and how they are unique
HBOBP:
Leghemoglobin= binds to O2 so that bacteria is able to do nitrogen fixation
chlorocruorin= green in annelids
NHBOBP:
found only in inverts
hemerythrin= 2 Fe and intracellular protein
hemocyanin= blue extracellular O2 transporter in mollusks and arthropods that binds to copper
Leghemoglobin= binds to O2 so that bacteria is able to do nitrogen fixation
chlorocruorin= green in annelids
NHBOBP:
found only in inverts
hemerythrin= 2 Fe and intracellular protein
hemocyanin= blue extracellular O2 transporter in mollusks and arthropods that binds to copper